
Data Balancing using Deep Convolutional Generative Adversarial
Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus
´
Erika G. Assis, Mark A. Song, Luis E. Z
´
arate and Cristiane N. Nobre
Department of Computing, Pontifical Catholic University of Minas Gerais University, Brazil
Keywords:
Congenital Syndrome, Zika, Generative Adversarial Networks, GAN, DCGAN.
Abstract:
Class imbalance is a common health care problem and often affects the performance of machine learning
algorithms. Unfortunately, the minority class, generally the one with the most significant interest, has their
learning affected to the detriment of the majority class. This article proposes using Deep Convolutional Gen-
erative Adversarial Networks (DCGAN) for minority class oversampling, generating synthetic instances. For
this, the ’RESP-Microcephaly’ database was used, which records suspected cases of congenital alteration due
to Zika virus (ZIKV) infection. The database presents unbalanced data with 2904 and 7606 instances with
and without congenital alteration, respectively. To evaluate the performance of DCGAN, we compared this
method with an undersampling and an oversampling approach, using SMOTE with three classification algo-
rithms. The use of DCGAN for balancing demonstrates a significant improvement in classification indices,
especially about the minority class.
1 INTRODUCTION
In recent years, machine learning techniques have
been applied to several domains, especially in the
health area such as breast cancer, thyroid disease,
Parkinson’s disease, predict mortality rate, and life
expectancy (Tomar, 2013) (Herland et al., 2014) (Jap-
kowicz, 2000a) (Batista et al., 2004) (Alsharqi et al.,
2018) (Weng et al., 2017) (Green, 2018) (Esteva et al.,
2017).
However, these healthcare datasets often suffer
from “rare class” issues, which result in unbalanced
classes in the training datasets (Batista et al., 2004)
(Chawla, 2005) (Milovic and Milovic, 2012). That is,
most health datasets generally have very few cases of
the target disease compared to the number of healthy
patients in the dataset (Chawla, 2005) (Japkowicz,
2000b). In the binary classification for medical diag-
nosis, the rare minority class refers to the positive in-
stances or target class. In contrast, the majority class
is represented by the negative cases in the dataset.
Although unbalanced data is frequent in machine
learning tasks, this is a very challenging task for clas-
sification algorithms (Chawla et al., 2003). This is be-
cause traditional machine learning methods applied to
unbalanced problems usually have a bias in favor of
the majority class, with unsatisfactory performance
in the minority class. This takes place during train-
ing; the minority classes collaborate less towards the
minimization of the objective function (Chawla et al.,
2002).
There are traditional techniques for working with
class imbalance. One approach is subsampling, which
involves excluding majority class instances to balance
instances in each class. Unfortunately, despite repair-
ing the disproportion, the classifier loses the majority
of information and is also likely to make sampling er-
rors on small data sets (Japkowicz, 2000a). Another
technique is oversampling, in which instances of the
minority class are augmented so that the classes are
uniform. This solves the balance problem but may
cause the classifier to generalize less to the minority
class because the particulars of the minority class data
become more usual (Japkowicz, 2000a).
An example of a widely used algorithm that uses
this approach is the SMOTE (Chawla et al., 2002)
(Chawla et al., 2003). It increases the number of
instances of the minority class by creating synthetic
examples. First, a sample belonging to the minority
class is randomly selected to generate new instances,
considering its neighbors. Then, from this neighbor-
hood, a new sample is built with the interpolation of
neighboring points. For this, lines are drawn between
the examples that make up the neighborhood. On
these lines, synthetic points belonging to the minority
class are generated (Chawla et al., 2002). Thus, al-
Assis, É., Song, M., Zárate, L. and Nobre, C.
Data Balancing using Deep Convolutional Generative Adversarial Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus.
DOI: 10.5220/0010842900003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 93-102
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
93

though these new instances may not accurately reflect
the actual distribution of the data, they tend to be close
enough to encourage generalization and increase the
accuracy of the overall classification (Chawla et al.,
2002). On the other hand, deep learning methods have
been used very efficiently, as is the case of Generative
Adversarial Networks (GANs) (Mariani et al., 2018)
(Mullick et al., 2019).
The GANs were inspired by game theory; the gen-
erator (G) and the discriminator (D) complement each
other until reaching the Nash equilibrium in the train-
ing process (Goodfellow et al., 2014). Both are neural
networks with different responsibilities. The discrim-
inator is a network responsible for evaluating whether
certain content is real or generated. The generator
produces the content itself. The relationship between
these two components is performed in an adversarial
way. At the same time, the discriminator is enabled
to distinguish the real from the fake, and the genera-
tor is trained to deceive the discriminator through the
content it is producing. Through the training of both,
networks develop together to determine their respon-
sibilities (Goodfellow et al., 2014).
This article uses a synthetic oversampling ap-
proach called DCGAN (Deep Convolutional Genera-
tive Adversarial Network), a GAN type that explicitly
uses convolutional and convolutional transpose lay-
ers in the discriminator and generator (Salimans et al.,
2016).
Generating artificial data through data augmenta-
tion (DA) techniques can be an alternative to improve
classification. Several works have already applied DA
and obtained improvement in their results (Hussain
et al., 2018), (Wang et al., 2017) and (Yu et al., 2017).
We used GAN’s to perform AD and tested it on a
set of tabular data from the Public Health Event Reg-
istry RESP-Microcephaly
1
, which presents an imbal-
ance of classes in the order of 1:2.6. There is a ma-
jority class with 72.37% (without syndrome), much
more frequently than the minority class (with the syn-
drome) 27.63%
In addition to the two oversampling balancing
methods, SMOTE and GANs, we also compared the
Random Under Sampler (RUS). The RUS undersam-
pling method removes the majority class samples at
random; in the end, the majority class has the same
number of samples as the minority class.
1
The RESP-Microcephaly is an online form devel-
oped by DATASUS-Brazil, instituted by the Ministry of
Health (MS), since November 19, 2015, to record cases
and deaths suspected of changes in growth and devel-
opment related to infection by the Zika virus and other
infectious etiologies (Brasil et al., 2015). Available at:
http://www.resp.saude.gov.br/microcefalia
Thus, this work aims to investigate data balancing
methods in diagnosing newborns and children with
congenital syndrome caused by ZIKV infection. Re-
garding classifiers, we use three algorithms: Random
Forest, Decision Tree, and Bagging.
This work is structured as follows: Section 2
brings the background used in the research. Section
3 presents the works related to the topic investigated.
Section 4 describes the materials and methods used in
the experiments. Finally, in Section 5, the results and
Discussions, and Section 6 presents the final consid-
erations and proposals for future work.
2 BACKGROUND
2.1 Congenital Zika Syndrome
On March 31, 2016, the World Health Organization
(WHO) announced Zika virus infection (ZIKV) as an
emergency public health problem worldwide due to
the association of this arbovirus with the occurrence
of congenital Zika syndrome.
ZIKV is mainly transmitted by the vector Aedes
aegypti, which resides in tropical and subtropical re-
gions, as well as by Aedes albopictus, the inhabi-
tant of the European Mediterranean (Carvalho et al.,
2019).
Mothers can transmit the Zika virus to embryos
or fetuses during pregnancy or at birth time (Zanluca
et al., 2017).
Children born to women infected with ZIKV dur-
ing pregnancy showing varying degrees of nervous
system impairment, such as microcephaly and other
neurodevelopmental lesions (Boeuf et al., 2016).
In additional observational studies, a set of con-
genital anomalies was identified and linked to ZIKV
infection in the uterus, called Congenital Zika Syn-
drome (CZS). This syndrome includes, in addition to
microcephaly, craniofacial disproportion, irritability,
spasticity, seizures, feeding difficulties, visual abnor-
malities, and hearing loss, as well as calcifications,
cortical disorders, and fetal cerebral ventricle dilata-
tion (Lima et al., 2019).
This article aims at improving CZS classifica-
tion by balancing GAN’s by improving classification
methods to improve early diagnosis and prevention.
2.2 Generative Adversarial
Networks - GAN
The Adversary Generative Networks (GANs), pro-
posed by (Goodfellow et al., 2014), are deep neu-
HEALTHINF 2022 - 15th International Conference on Health Informatics
94

ral network architectures composed of two networks
placed against each other. The authors call this model
adversary networks and training both models using
only the backpropagation and dropout algorithms, be-
ing highly successful.
GANs are a kind of differentiable generator net-
work, which is, we can use backpropagation to train
with a descending gradient (Goodfellow et al., 2014).
This type of model transforms samples from a latent
vector z into examples x using the smooth function
g(z, θ) (Goodfellow et al., 2014). Essentially, dif-
ferentiable generator networks are computing proce-
dures for generating samples.
A typical architecture for GANs is illustrated in
Figure 1. The generator is an G differentiable func-
tion. When z is sampled from some previous simple
distribution, G(z) yields a sample of x (Goodfellow
et al., 2014).
The generator input is a random noise vector Z,
usually a uniform or normal distribution. The noise is
mapped to a new data space via the G generator to ob-
tain a false sample, G(z), which is a multidimensional
vector (Goodfellow et al., 2014) (Pan et al., 2019).
The generator is G differentiable function. When
z is sampled from some previous simple distribu-
tion, G(z) produces a sample of x (Goodfellow et al.,
2014).
The generative network tries to produce samples
that resemble the original data, x yesP
d
ata, according
to the series of transformations that can be described
by the function x = g(z;θ
g
).
The D discriminating is a binary classifier. It takes
an accurate sample from the dataset. The false piece
generated by the G generator as input and the out-
put from the D discriminator represents the probabil-
ity that the example is authentic.
The discriminating network, in turn, produces the
likelihood of x being false or real, which is given by a
function d(x; θ
d
). D is trained to maximize the proba-
bility of a hit of a sample being genuine or false (com-
ing from G), and while G is trained to minimize the
likelihood of being discovered, [log(1 − D(G)(z))].
In practice, Goodfellow et al. (2014) have ob-
served that minimize [log(1 − D(G)(z))] makes gra-
dients converge to zero quickly and maximizing
log(D(G(z))) is equivalent and allows for gradients
with higher values. Consequently, the optimization
problem of GANs is transformed into the objective
function presented in Equation 1.
min
G
∼ max
D
V (D, G) =
= E
x∼data
(x)[logD(x)] + E
z∼pz(z)
[log(1 − D(G)(z))],
(1)
So the game between Generator and Discrimina-
tor is established, the function V (D, G) will reach
its maximum value when the Nash equilibrium is
reached, that is, when neither of the two can improve
its performance. Thus, the above function will be
used as the trouble loss function.
Thus, in its simplest form, given the two play-
ers (discriminator and generator), the GAN’s learning
problem is solved as a zero-sum game, when the gain
obtained by one participant is equivalent to the loss
by the other participant, where the function r(θ
d
;θ
g
)
determines the reward for one of the networks and
−r(θ
d
;θ
g
) for the other.
As the game is zero-sum, in Nash’s unique equi-
librium you will have:
g
∗
= arg min
g
max
d
r(g, d) (2)
where g
∗
is the generating network at the conver-
gence point, optimally capturing the data distribution.
The standard choice for r is
r(θ
d
;θ
g
) = E
x
∼ P
data
log(d(x)) + E
x
∼ P
model
[log(1 − d(x))]
(3)
We can separate the cost function of each one of
the networks, for the discriminator we would have
L
g
=
−1
m
∑
[log(d(x))] + [log(1 − d(g)(z))] (4)
The second term, log(1 − d(g)(z)), concerns the
incorrect classification of false samples. The equa-
tion 5 gives the cost to be optimized by the network
generator.
L
g
=
1
m
∑
[log(1 − d(g)(z))] (5)
Intuitively, these cost functions make it the objec-
tive of the discriminating network to maximize the
correctness of the classification of samples into false
and accurate; the generating network will try to mini-
mize these hits.
3 RELATED WORKS
Adversary Generative Networks (GANs) have cur-
rently been used in machine learning problems in
unbalanced (Japkowicz, 2000a) databases. We will
present below the main works that used GAN’s for
oversampling of the minority class in several areas of
knowledge.
Mehta et al. (2019) used GAN’s to help improve
the images of those who suffered a stroke. Han et al.
(2019) investigated magnetic resonance (MR) images
Data Balancing using Deep Convolutional Generative Adversarial Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus
95
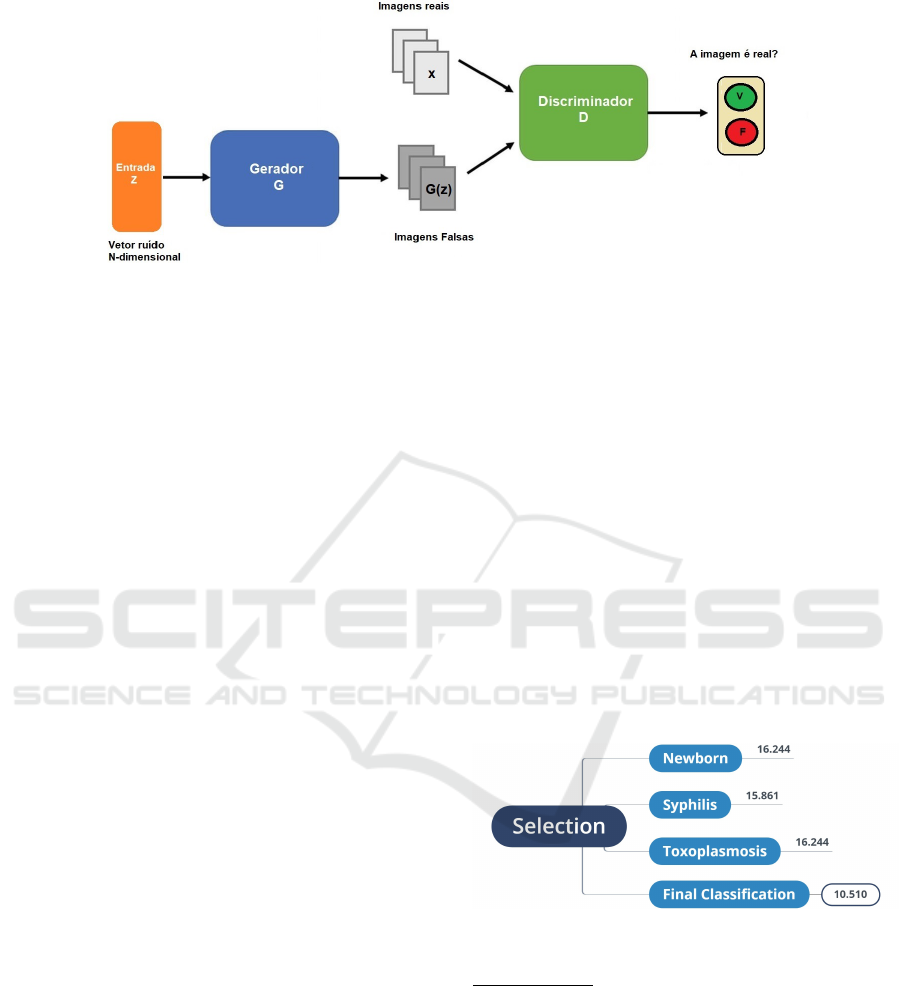
Figure 1: GANs architecture. Source: Based on (Goodfellow et al., 2014).
for tumor detection. Bhagat and Bhaumik (2019) cre-
ated synthetic chest X-ray images of pneumonia pa-
tients to improve the accuracy of the image classi-
fication. Bailo et al. (2019) augmented the blood
smear microscopic image datasets, which are im-
ages of blood taken with a microscope, where a thin
layer of blood is placed on a microscope slide (Bailo
et al., 2019). Asvestopoulou et al. (2019) augmented
the speech dataset to improve DysLexML’s dyslexia
screening tool ranking. Sheng et al. (2019) created
datasets to improve speech recognition in children un-
der noisy conditions (Hu et al., 2018). (Haradal et al.,
2018) and (Fahimi et al., 2020) increased the biosig-
nal dataset (electrocardiogram and electroencephalo-
gram) to improve ranking.
All works obtained promising experimental re-
sults, even with few original data, implying that the
generated data can be used as extended samples to in-
crease a database to improve classification tasks in the
most diverse applications.
It was also observed that GAN performance is
sensitive to model parameters such as learning rate,
number of epochs, and others (Karadag and Erdas¸ Ci-
cek, 2019).
Data augmentation with GANs is sensitive to the
size of the dataset because when the number of im-
ages increases to more than 60,000, the synthetic im-
ages generated by the GAN do not contribute to the
classification performance and may even cause a re-
duction in performance (Karadag and Erdas¸ Cicek,
2019).
4 MATERIALS AND METHODS
This section presents the materials and methods used
in this work. As well as an overview of the RESP-
Microcephaly database that records suspected cases
of genetic alteration due to Zika virus (ZIKV) infec-
tion. A descriptive analysis of the database is also
presented, in addition to its pre-processing.
4.1 RESP Database
Initially, the database had 17451 instances. As the in-
terest of the work is to classify children born with ge-
netic alterations due to Zika virus infection, we work
with the notifications of newborns and children
2
. We
also excluded all subjects with laboratory confirma-
tion for syphilis or toxoplasmosis
3
.
The instance selection process summarized in Fig-
ure ref Selection. Initially, there were 14,144 cases
and confirmed cases with syphilis and toxoplasmosis
excluded. We only work with congenital changes due
to ZKV and not other causes. At the end of the se-
lection process, we have 10,510 instances, 2,904 chil-
dren with genetic alterations, and 7,606 children with-
out alterations.
Figure 2: Selection of instances from the RESP.
2
These children already had Microcephaly before the
Zika outbreak and were included in the RESP under the
guidance of the Secretary of Health to follow up on cases
(Brasil et al., 2015).
3
Microcephaly may be associated with various envi-
ronmental and genetic factors. Among the environmental
factors there is fetal distress, congenital STORCH infec-
tions. The acronym is composed of the pathogens most fre-
quently related to diseases: Treponema Pallidum bacteria
that causes syphilis (S), the protozoan Toxoplasma Gondii
that causes toxoplasmosis (TO) and the rubella virus (R),
cytomegalovirus (C), herpes virus simple (H) (Ribeiro et al.,
2018).
HEALTHINF 2022 - 15th International Conference on Health Informatics
96

Thus, the RESP database had 10.510 instances
and 43 attributes, organized into nine (9) categories:
1. Notification: Displays the classification of sus-
pected cases of congenital infection (newborn,
child, fetus at risk, miscarriage, or stillbirth) and
the date it notified
2. Pregnant Woman’s Data: age, race/color, and
state of residence (UF)
3. Information about Live Births: sex, date of birth,
weight (grams), and length (centimeters)
4. Data on Pregnancy and Childbirth: types of con-
genital changes, when the change was detected (in
pregnancy or after delivery), gestational age at de-
tection of microcephaly, type of pregnancy, classi-
fication of live birth, head circumference, and date
of head circumference measurement. The type of
pregnancy that defined as preterm (gestational age
less than 37 weeks of gestation), the term (gesta-
tional age between 37 and 41 weeks of gestation),
post-term (gestational age greater than 42 weeks)
5. Mother’s Clinical, Epidemiological Data: date of
onset of symptoms, type of symptoms (fever, rash,
itching, conjunctivitis, headache, and neurologi-
cal involvement), Syphilis/Toxoplasmosis test and
result, Zika test results, history of arboviruses, and
congenital malformations
6. Information about Imaging Tests: ultrasound,
transfontanellar ultrasound, computed tomogra-
phy, and magnetic resonance
7. Data about the Health Establishment: municipal-
ity and state
8. Data on Disease Evolution: death and date of
death
9. Fields Restricted to the Manager: Final classi-
fication of the suspected case of Congenital al-
terations and Confirmation criteria through lab-
oratory tests performed (Zika, Dengue, Chikun-
gunya, Syphilis, and Toxoplasmosis, others and
image)
These categories, together with their attributes, are
shown in Figure 3.
The occurrences registered in the RESP confirmed
congenital ZIKV infection peaked in 2016 with more
than 1600 records. As of May 2016, there is a drop
in the number of cases, a behavior observed in subse-
quent years, as shown in Figure 4.
The cases are distributed throughout the Brazilian
territory, as shown in Figure 5. The ten states that
presented the highest number of positive diagnoses
were: Bahia, Pernambuco, Rio de Janeiro, Para
´
ıba,
Maranh
˜
ao, Cear
´
a, Sergipe with, respectively: 490,
452, 259, 193, 166, 146, 134 cases; in addition to the
states of Alagoas and Rio Grande do Norte, both with
130 cases.
About the pregnant women’s region, 60.8% of the
records are from the Northeast region, 24.9% from
the Southeast, 6.7% from the Midwest, 4.3% from
the North region, and only 3.3% from the Southern
region.
4.2 Preprocessing
Before applying the classification algorithms effec-
tively, simple pre-processing strategies were adopted
to obtain a more consistent and impartial model. The
database processing phases were:
• Attribute Binarization: The original RESP was
composed of numerical (13%) and categorical
(87%) attributes. We perform one-hot coding to
binarize all categorical attributes. At the end of
the process, the database had 56 attributes.
• Inconsistent Data: There were 95 instances in
which the pregnant woman’s age was with values
2 and 3. As this is a physiologically incompati-
ble age for a conception, we excluded these values
and left them blank.
Two instances had the brain circumference value
measuring 323.3 cm, not corresponding to an ac-
tual value. These literature reports refer to a
mean head circumference of 34.61 cm in typi-
cal male NBs, ranging between 32.14 and 37.08
cm, and an average of 34.05 cm in normal female
NBs. with variation between 31.58 and 36.52 cm
(Brasil et al., 2015). As these values do not cor-
respond to values found in the literature, we ex-
cluded these values, leaving them absent.
• Missing Data: Missing data is common in health
databases. Therefore, the use of proper methods
becomes essential to reduce the impact of infor-
mation loss. The original database was about 30
% missing data. We deal with missing data by
imputing the data via the mean and median.
• Sampling Methods: There are two types of sam-
pling methods: undersampling and oversampling.
Undersampling removes elements from the major-
ity class while oversampling seeks to include ele-
ments from the minority class.
Random oversampling was implemented using the
RandomOverSampler class in Python. The class used
the sampling strategy argument set to “minority” to
balance the minority class with the majority class au-
tomatically.
Data Balancing using Deep Convolutional Generative Adversarial Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus
97
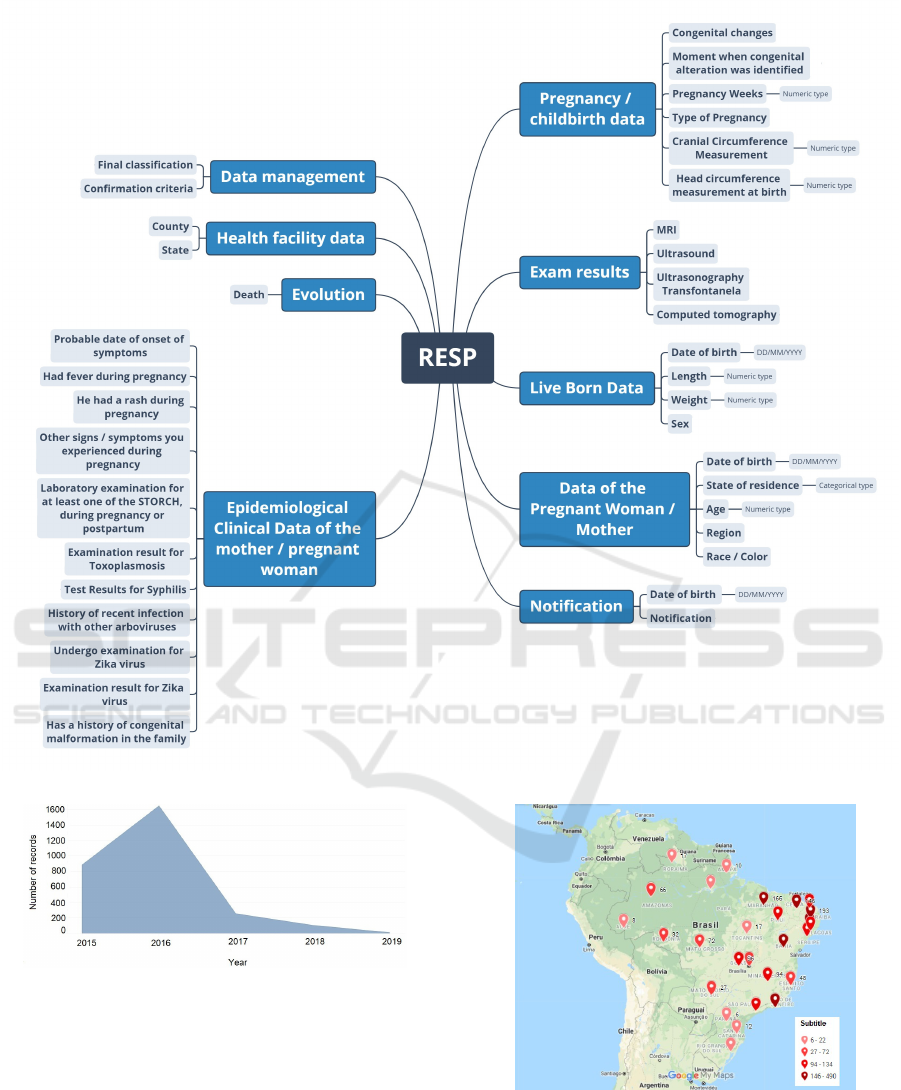
Figure 3: Categories and their respective attributes Registration of Public Health Events - RESP.
Figure 4: Confirmed cases of congenital infection due to
Zika Virus in Brazil between 2015 and 2019.
In work we use two oversampling methods: 1)
SMOTE (Synthetic Minority Over-sampling TEch-
nique), which generates synthetic cases for the class
of interest from existing data. The new data are gen-
erated in the neighborhood of the minority class data
to increase the decision space of this class and in-
crease the generalization power of the obtained clas-
sifiers (Chawla et al., 2002); 2) GAN, in which the ar-
chitecture used was a DCGAN (Deep Convolutional
Generative Adversarial Network) that allows training
Figure 5: Brazil - Cases of congenital infection due to Zika
Virus.
a pair of deep convolutional networks: generator and
discriminator.
DCGAN is a GAN model that uses deconvolution
layers in the generator and convolution layers in the
discriminator to extract characteristics from the data
HEALTHINF 2022 - 15th International Conference on Health Informatics
98

and build a model to generate the synthetic data.
The first layer of the generator receives an evenly
distributed N-dimensional noise as an input to a fully
connected network. Then, the result is remodeled in a
series of four convolutions with fractional steps (512,
256, 128, 64), according to Figure 6.
DCGAN combines the deep learning stage as the
key to GAN training. These techniques include the
fully convolutional network and Batch Normalization
(BN). Batch normalization is a technique initially in-
troduced by (Ioffe and Szegedy, 2015). Batch nor-
malization is a solution to speed up the training phase
of deep neural networks by introducing internal nor-
malization of input values in the neural network layer.
The first emphasizes magnified convolutions
(rather than grouped layers) for both: increasing and
decreasing the spatial dimensions of the feature. Sec-
ond, normalizes feature vectors to have zero mean and
unity variance across all layers, helping to stabilize
learning and handle underweight startup problems.
The generator network has four convolutional lay-
ers. All followed by BN (except for the output layer)
and rectified linear activation (ReLU). In addition, the
generator receives as input a random z vector (ob-
tained from a normal distribution).
The discriminator is also a 4-layer CNN with BN
(except its input layer) and leaky RELU triggers.
Many enablement functions will work fine with this
basic GAN architecture. However, leaky ReLUs are
very popular because they help gradients to flow more
easily across the architecture.
A regular ReLU function works by truncating neg-
ative values to zero, blocking the flow of gradients
across the network. However, instead of the func-
tion being zero, leaking RELUs allow a small nega-
tive value to pass. That is, the function calculates an
immense value between resources and a smaller fac-
tor.
The generator output corresponds to the 55 re-
sources of the dataset, plus an extra neuron to enable
class discrimination. In the discriminator, the final
convolution layer is flattened and then fed to a single
sigmoid output.
4.3 Assessment Metrics
In this work, the following performance evaluation
measures were used: precision, recall and F-Score.
Precision (Equation 6) identifies, among all in-
stances classified in a given class, those that are ac-
tually of the class in question.
Precision =
T P
T P + FP
(6)
wich: TP = True positive, TN = True Negative, FP =
False positive, and FN= False Negative.
Recall (Equation 7) measures the hits in a given
class. That is, among all the instances of a given class,
how many actually the classifier classified as being of
the class.
Recall =
T P
T P + FN
(7)
F-Score is a harmonic mean between precision
and recall and indicates the overall quality of the
model, given by Equation 8.
F − Score =
2 ∗ precision ∗ recall
precision + recall
(8)
5 RESULTS AND DISCUSSIONS
To evaluate the performance of using GANs, we
compared this method with two traditional balancing
methods: Undersampling and SMOTE, and with the
results with unbalanced classes.
We split the dataset into 80% for model creation
and 20% for testing. 10-fold cross-validation was
used to create the models.
The unbalanced dataset had 2904 instances of the
class “yes” and 7606 instances of class “no”. In
the sub-sampling, the data of the majority class were
reduced so that, in the end, the data set was com-
posed of 2336 instances of each class. To avoid a bi-
ased model, the oversampling method was applied in
cross-validation. In other words, for every nine train-
ing folds, the oversampling method was used.
In addition, we use three learning algorithms:
Bagging, Random Forest, and Decision Tree. The re-
sults obtained are shown in Figure 7.
Analyzing the results, we see that the highest pre-
cision for the ‘Yes’ class was 94% with data balancing
with GAN using the Random Forest classifier. This
means that only 6% of the data were classified as false
positives, that is, individuals who were identified as
having the congenital syndrome but actually did not
have the alteration.
Regarding the precision for class ‘No,’ the best re-
sults were also balanced with GAN in which the three
classifiers had the same 90% performance; this repre-
sents that 10% of the instances were classified as not
having the syndrome, but actually they were.
Regarding the Recall rate, the best index for the
‘Yes’ class was 90% for balanced data with GAN with
Random Forest. This result represents that 10% of the
patients were classified as not having a congenital al-
teration and were carriers. For the ‘No’ class, the best
Data Balancing using Deep Convolutional Generative Adversarial Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus
99
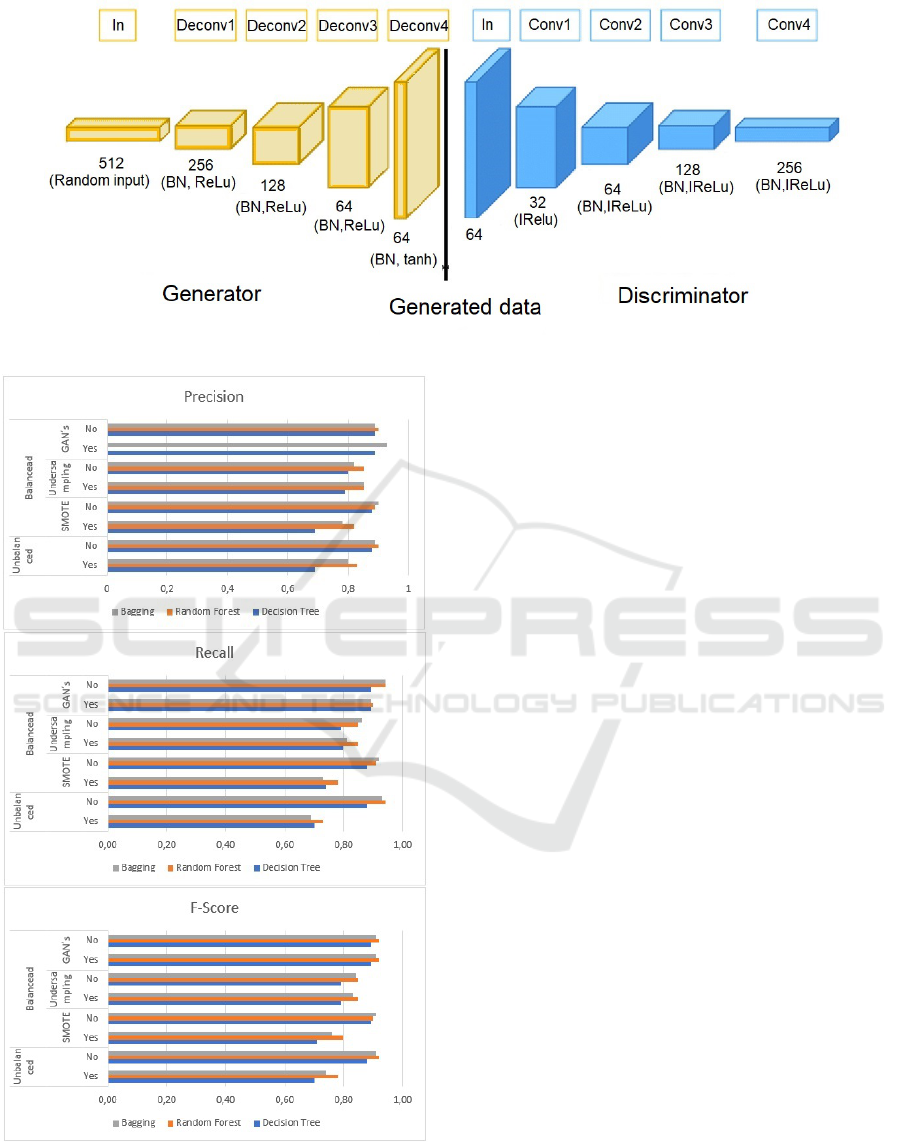
Figure 6: Structure of the deep convolutional generator for DCGAN.
Figure 7: Classification metrics with balanced and unbal-
anced datasets.
rate was 94% for unbalanced and balanced data with
GAN, for Bagging and Random Forest. This means
that 6% were false negatives; that is, individuals clas-
sified as having congenital alterations, but on the con-
trary, they didn’t have it.
For the F-Score metric, for both classes, the best
rate was 92% in balancing with GAN using Random
Forest classifier.
Regarding the unbalanced data, we found that
the algorithms ranked the majority class better (not),
which was expected to happen, as there is a significant
difference between the classes.
When we work with SMOTE, there are a slight
improvement in the “yes” class results. When we
compare the ”no” class results, all metrics improve
for all algorithms.
Therefore, we can conclude that the undersam-
pling method significantly improves the classifica-
tion of the ‘yes’ minority sample, which is expected
since we took samples from the majority class. With
SMOTE, there is a slight improvement for the minor-
ity class, but it continues to rank the majority class
better. Finally, with GAN’s there was a significant
gain in all metrics for both classes.
It is noted that balancing data with GAN showed
a significant improvement in the indices, especially
concerning the minority class ‘Yes,’ as with the cre-
ation of synthetic data with GAN, the minority class
gains more importance, and the bias on the class ma-
jority is slight.
6 FINAL CONSIDERATIONS
This work uses Adverse Generative Networks to syn-
thesize data to oversample minority classes in unbal-
anced datasets and compares the results with other
balancing algorithms. The results suggest that GAN
HEALTHINF 2022 - 15th International Conference on Health Informatics
100

can increase the classifier performance for all evalu-
ated metrics since, in all metrics (accuracy, recall, and
F-Score), for all classification algorithms, the values
were above 90%.
Observed that there is a significant improvement,
especially about the minority class, as with the cre-
ation of synthetic data, there was an increase in the
representation and density of the data.
As most classification models are designed to
work with balanced datasets, GANs for data balanc-
ing add a greater generalization power of the algo-
rithms, detecting rare and essential patterns that dis-
criminate the classes of the problem by establishing a
reliable decision threshold.
Another point worth mentioning is that data on
CZS are rare data since, since 2019, the Federal
Government has considered the data as confidential
and no longer makes this information available to re-
searchers and the general public
4
.
Therefore, this detailed analysis of this dataset
and, above all, the significant improvement in the
classification process, add the importance of using
GANs for balancing tabular datasets and, above all,
for generating synthetic data from rare and restricted
data as it is our case.
Furthermore, the approach presented in this arti-
cle has the potential for early diagnosis of congenital
syndrome associated with Zika virus infection. Early
diagnosis increases prevention, speeds up treatment,
and reduces the devastating consequences of this ill-
ness for mothers and children.
In future works, we suggest the refinement of
the model proposing the application of deep learning
techniques to the GAN architecture to deal with dif-
ferent data types and performing statistical analysis
and uncertainty analysis of the results obtained.
ACKNOWLEDGEMENTS
The authors thank the National Council for Scientific
and Technological Development of Brazil (CNPq),
the Coordination for the Improvement of Higher Ed-
ucation Personnel - Brazil (CAPES), the Founda-
tion for Research Support of Minas Gerais State
(FAPEMIG) and Pontifical Catholic University of Mi-
nas Gerais (PUC Minas).
4
The report is available in a way that has al-
ready been processed by state and is available at:
https://datasus.saude.gov.br/acesso-a-informacao/registro-
de-eventos-em-saude-publica-resp-microcefalia/
REFERENCES
Alsharqi, M., Woodward, W., Mumith, J., Markham, D.,
Upton, R., and Leeson, P. (2018). Artificial intelli-
gence and echocardiography. Echo research and prac-
tice, 5(4):R115—R125.
Asvestopoulou, T., Manousaki, V., Psistakis, A., Nikolli,
E., Andreadakis, V., Aslanides, I., Pantazis, Y., Smyr-
nakis, I., and Papadopouli, M. (2019). Towards a
robust and accurate screening tool for dyslexia with
data augmentation using gans. In 2019 IEEE 19ª Con-
fer
ˆ
encia Internacional sobre Bioinform
´
atica e Bioen-
genharia (BIBE), pages 775–782.
Bailo, O., Ham, D., and Shin, Y. (2019). Red blood cell
image generation for data augmentation using condi-
tional generative adversarial networks. In 2019 IEEE
/ CVF Conference on Computer Vision and Pattern
Recognition Workshops (CVPRW), pages 1039–1048.
Batista, G. E. A. P. A., Prati, R., and Monard, M. C. (2004).
A study of the behavior of several methods for balanc-
ing machine learning training data. SIGKDD Explor.,
6:20–29.
Bhagat, V. and Bhaumik, S. (2019). Data augmentation
using generative adversarial networks for pneumonia
classification in chest xrays. In 2019 Fifth Interna-
tional Conference on Image Information Processing
(ICIIP), pages 574–579, Shimla, India, 2019.
Boeuf, P., Drummer, H., Richards, J., Scoullar, M., and
Beeson, J. (2016). The global threat of zika virus
to pregnancy: Epidemiology, clinical perspectives,
mechanisms, and impact. BMC Medicine, 14:112.
Brasil, da Sa
´
ude, M., de Vigil
ˆ
ancia em Sa
´
ude, S., and
de Vigil
ˆ
ancia das Doenc¸as Transmiss
´
ıveis., D. (2015).
Protocolo de vigil
ˆ
ancia e resposta
`
a ocorr
ˆ
encia de mi-
crocefalia e/ou alterac¸
˜
oes do sistema nervoso central
(snc): emerg
ˆ
encia de sa
´
ude p
´
ublica de import
ˆ
ancia in-
ternacional.
Carvalho, I. F., Alencar, P. N. B., Carvalho de Andrade,
M. D., Silva, P. G. d. B., Carvalho, E. D. F., Ara
´
ujo,
L. S., Cavalcante, M. P. M., and Sousa, F. B. (2019).
Clinical and x-ray oral evaluation in patients with con-
genital Zika Virus. Journal of applied oral science :
revista FOB, 27:e20180276–e20180276.
Chawla, N. (2005). Data Mining for Imbalanced Datasets:
An Overview, volume 5, pages 853–867. Springe.
Chawla, N., Japkowicz, N., and Kolcz, A. (2003). Work-
shop learning from imbalanced data sets ii. In
Proceedings of international conference on machine
learning.
Chawla, N. V., Bowyer, K. W., Hall, L. O., and Kegelmeyer,
W. P. (2002). Smote: Synthetic minority over-
sampling technique. Jornal de pesquisa de in-
telig
ˆ
encia artificial, 16:321–357.
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M.,
Blau, H. M., and Thrun, S. (2017). Dermatologist-
level classification of skin cancer with deep neural net-
works. Nature, 542(7639):115—118.
Fahimi, F., Dosen, S., Ang, K. K., Mrachacz-Kersting,
N., and Guan, C. (2020). Generative adversarial
networks-based data augmentation for brain-computer
Data Balancing using Deep Convolutional Generative Adversarial Networks (DCGAN) in Patients with Congenital Syndrome by Zika Virus
101

interface. IEEE Transactions on Neural Networks and
Learning Systems, pages 1–13.
Goodfellow, I. J., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. In
Proceedings of the 27th International Conference on
Neural Information Processing Systems - Volume 2,
NIPS’14, page 2672–2680, Cambridge, MA, USA.
MIT Press.
Green, M. A. (2018). Use of machine learning approaches
to compare the contribution of different types of data
for predicting an individual’s risk of ill health: an ob-
servational study. The Lancet, 392:p.S40.
Han, C., Murao, K., Noguchi, T., Kawata, Y., Uchiyama,
F., Rundo, L., Nakayama, H., and Satoh, S. (2019).
Learning more with less: Conditional pggan-based
data augmentation for brain metastases detection us-
ing highly-rough annotation on mr images. In Pro-
ceedings of the 28th ACM International Conference
on Information and Knowledge Management, CIKM
’19, page 119–127, New York, NY, USA. Association
for Computing Machinery.
Haradal, S., Hayashi, H., and Uchida, S. (2018). Biosig-
nal data augmentation based on generative adversarial
networks. In 2018 40th Annual International Confer-
ence of the IEEE Engineering in Medicine and Biol-
ogy Society (EMBC), pages 368–371.
Herland, M., Khoshgoftaar, T., and Wald, R. (2014). A re-
view of data mining using big data in health informat-
ics. Journal Of Big Data, 1:2.
Hu, H., Tan, T., and Qian, Y. (2018). Generative adversar-
ial networks based data augmentation for noise robust
speech recognition. In 2018 IEEE International Con-
ference on Acoustics, Speech and Signal Processing
(ICASSP).
Hussain, Z., Gimenez, F., Yi, D., and Rubin, D. (2018).
Differential data augmentation techniques for medical
imaging classification tasks. AMIA ... Annual Sympo-
sium proceedings. AMIA Symposium, 2017:979–984.
Ioffe, S. and Szegedy, C. (2015). Batch normalization:
Accelerating deep network training by reducing in-
ternal covariate shift. In Bach, F. and Blei, D., ed-
itors, Proceedings of the 32nd International Confer-
ence on Machine Learning, volume 37 of Proceedings
of Machine Learning Research, pages 448–456, Lille,
France. PMLR.
Japkowicz, N. (2000a). The class imbalance problem: Sig-
nificance and strategies. In Proc. of the Int’l Conf. on
Artificial Intelligence, volume 56. Citeseer.
Japkowicz, N. (2000b). The class imbalance problem: Sig-
nificance and strategies. Proceedings of the 2000 In-
ternational Conference on Artificial Intelligence ICAI.
Karadag, O. O. and Erdas¸ Cicek, O. (2019). Experimen-
tal assessment of the performance of data augmenta-
tion with generative adversarial networks in the image
classification problem. In 2019 Innovations in Intel-
ligent Systems and Applications Conference (ASYU),
pages 1–4.
Lima, G. P., Rozenbaum, D., Pimentel, C., Frota, A. C. C.,
Vivacqua, D., Machado, E. S., das Neves Sztajnbok,
F. C., Abreu, T., Soares, R. A., and Hofer, C. B.
(2019). Factors associated with the development of
congenital zika syndrome: a case-control study. In
BMC Infectious Diseases.
Mariani, G., Scheidegger, F., Istrate, R., Bekas, C., and
Malossi, A. C. I. (2018). Bagan: Data augmentation
with balancing gan. arXiv, pages 1–9.
Mehta, K., Kobti, Z., Pfaff, K., and Fox, S. (2019). Data
augmentation using ca evolved gans. 2019 IEEE Sym-
posium on Computers and Communications (ISCC),
pages 1087–1092.
Milovic, B. and Milovic, M. (2012). Prediction and decision
making in health care using data mining. International
Journal of Public Health Science (IJPHS), 1.
Mullick, S. S., Datta, S., and Das, S. (2019). Generative ad-
versarial minority oversampling. In 2019 IEEE/CVF
International Conference on Computer Vision (ICCV),
pages 1695–1704.
Pan, Z., Yu, W., Yi, X., Khan, A., Yuan, F., and Zheng,
Y. (2019). Recent progress on generative adversarial
networks (gans): A survey. IEEE Access, 7:36322–
36333.
Ribeiro, I. G., Andrade, M. R. d., Silva, J. d. M. S., Silva,
Z. M., Costa, M. A. d. O., Vieira, M. A. d. C. e. S.,
Batista, F. M. d. A., Guimar
˜
aes, H., Wada, M. Y.,
and Saad, E. (2018). Microcefalia no Piau
´
ı, Brasil:
estudo descritivo durante a epidemia do v
´
ırus Zika,
2015-2016. Epidemiologia e Servic¸o de Sa
´
ude, 27.
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V.,
Radford, A., Chen, X., and Chen, X. (2016). Im-
proved techniques for training gans. In Lee, D.,
Sugiyama, M., Luxburg, U., Guyon, I., and Garnett,
R., editors, Advances in Neural Information Process-
ing Systems, volume 29. Curran Associates, Inc.
Sheng, P., Yang, Z., and Qian, Y. (2019). Gans for children:
A generative data augmentation strategy for children
speech recognition. In 2019 IEEE Automatic Speech
Recognition and Understanding Workshop (ASRU),
pages 129–135.
Tomar, D. (2013). A survey on data mining approaches for
healthcare. International Journal of Bio - Science and
Bio - Technology, 5:241–266.
Wang, S., Lv, Y.-D., Sui, Y., Liu, S., Wang, S.-J., and
Zhang, Y.-D. (2017). Alcoholism detection by data
augmentation and convolutional neural network with
stochastic pooling. Journal of Medical Systems, 42.
Weng, S. F. R., J.; Kai, J. G., and J. M.; Qureshi, N. (2017).
Can machinelearning improve cardiovascular risk pre-
diction using routine clinical data? Public Library of
Science, 12(4):e0174944.
Yu, X., Wu, X., Luo, C., and Ren, P. (2017). Deep learning
in remote sensing scene classification: a data augmen-
tation enhanced convolutional neural network frame-
work. GIScience & Remote Sensing, 54:1–18.
Zanluca, C., Noronha, L., and Santos, C. (2017). Maternal-
fetal transmission of the zika virus: An intriguing in-
terplay. Tissue Barriers, 6:00–00.
HEALTHINF 2022 - 15th International Conference on Health Informatics
102
