
Deep Features Extraction for Endoscopic Image Matching
Houda Chaabouni-Chouayakh
1,2
, Manel Farhat
1,2
and Achraf Ben-Hamadou
1,2
1
Centre de Recherche en Num
´
erique de Sfax, 3021, Sfax, Tunisia
2
Laboratory of Signals, Systems, Artificial Intelligence and Networks (SM@RT), Sfax, Tunisia
Keywords:
Endoscopic Images,Deep Learning, Image Feature Matching, Adaptive Triplet Loss.
Abstract:
Image feature matching is a key step in creating endoscopic mosaics of the bladder inner walls, which help
urologists in lesion detection and patient follow-up. Endoscopic images, on the other hand, are particularly
difficult to match because they are weekly textured and have limited surface area per frame. Deep learning
techniques have recently gained popularity in a variety of computer vision tasks. The ability of convolutional
neural networks (CNNs) to learn rich and optimal features contributes to the success of these methods. In this
paper, we present a novel deep learning based approach for endoscopic image matching. Instead of standard
handcrafted image descriptors, we designed a CNN to extract feature vector from local interest points. We pro-
pose an efficient approach to train our CNN without manually annotated data. We proposed an adaptive triplet
loss which has the advantage of improving the inter-class separability as well as the inter class compactness.
The training dataset is automatically constructed, each sample is a triplet of patches: an anchor, one positive
sample (a perspective transformation of the anchor) and one negative sample. The obtained experimental re-
sults show at the end of the training step a more discriminative space representation where the anchor becomes
closer to the positive sample and farther from the negative one in the embedding space. Comparison with the
well-known standard hand-crafted descriptor SIFT in terms of recall and precision showed the effectiveness
of the proposed approach, reaching the top recall value for a precision value of 0.97.
1 INTRODUCTION
Endoscopy is a widely used clinical procedure for the
early detection of numerous cancers (e.g., nasopha-
ryngeal, oesophageal adenocarcinoma, gastric, col-
orectal cancers, bladder cancer, etc.) and therapeutic
procedures (e.g., polypectomy, injection sclerother-
apy, variceal banding, etc.). An endoscope is the in-
strument used during an endoscopic exploration. It
could be a rigid or flexible thin tube equipped by a
lighting source and a camera. The doctor visualizes
the captured stream of video endoscopy while explor-
ing the inner surfaces of hollow organs.
Nevertheless, endoscopy has several limitations
due mainly to the limited field of view and reduced
maneuverability inside the bladder. Indeed, anoma-
lies are usually spread over areas larger than the field
of view making lesion partitions laid out over many
frames in the endoscopic video. This makes organs
exploration and anomalies detection time consuming
and difficult for the doctors since they should fol-
low the spatial distribution of the lesions over several
frames. In addition, the ability to find structure simi-
larities between images taken from different views or
during different clinical examinations is a tough task.
To address this issue, it is a common practice to re-
place the clinical endoscopic video with a panoramic
image generated by stitching the video frames in a
common coordinate system that covers a wide region
of interest. For the generation of panoramic images,
feature matching and finding correspondence between
images is a key step.
Because of the large variability in texture and
appearance aspect of the endoscopic images be-
tween patients, developing a robust and accurate fea-
ture matching between endoscopic images is partic-
ularly difficult. So far, this matching problem has
been addressed by either directly computing classi-
cal similarity measures between the images (e.g., mu-
tual information, cross-correlation, etc.) or by us-
ing hand-crafted features such as SIFT(Lowe, 2004),
SURF(Bay et al., 2008), etc. The main advantages
of such algorithms are the simplicity and the effi-
ciency. The basic idea in this case is to first ex-
tract features for a set detected image interest points.
After that, point matching between images is per-
Chaabouni-Chouayakh, H., Farhat, M. and Ben-Hamadou, A.
Deep Features Extraction for Endoscopic Image Matching.
DOI: 10.5220/0010833700003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
925-932
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
925

formed based on the distance between their feature
vectors. This category of techniques has previously
been used to stitch images for various applications in-
cluding remote sensing (Ait-Aoudia et al., 2012), un-
derwater navigation (Elibol et al., 2017), retina anal-
ysis (Hossein-Nejad and Nasri, 2018), and endoscopy
(Behrens et al., 2009; Bergen et al., 2013). In the case
of endoscopic images, we can cite for instance the
works of (Weibel et al., 2010; Ben-Hamadou et al.,
2016; Daul et al., 2010; Behrens et al., 2010; Du
et al., 2011). In particular, (Behrens et al., 2010) and
(Du et al., 2011) propose respectively a SURF fea-
ture descriptor for real-time bladder mosaicing in flu-
orescence endoscopy and a SIFT-based descriptor for
endoscopic zone matching. Recently, feature extrac-
tion from endoscopic images has been addressed in
the work of (Martinez et al., 2020) where color and
texture features are extracted and then exploited by
classical classifiers such as a random forest tree or a
KNN ensemble model for the classification of kidney
stone captured with ureteroscopes. Later, the authors
showed in (Lopez et al., 2021) how CNN-based solu-
tions can further improve the classification of kidney
stones, even when using a moderate number of im-
ages.
Nowadays, deep learning techniques clearly out-
perform standard and hand-crafted techniques for a
wide range of computer vision tasks including face
recognition (Lai et al., 2019), artifacts classification
(Ali et al., 2021), image feature matching (Liu et al.,
2018), denoising (Zou et al., 2019), classification
(Kim et al., 2021; Liu et al., 2020), and segmentation
(Ghosh et al., 2018). When it comes to endoscopy, to
the best of our knowledge, no previous study has been
proposed to build local features for endoscopic image
matching based on deep learning techniques.
In this context, we propose in this paper a new
deep learning-based method for learning local fea-
tures to improve endoscopic image matching. We de-
signed a CNN to transform local patches around im-
age interest points instead of applying standard hand-
crafted descriptors. The proposed training scheme
of the CNN does not require manual annotation of
data. We rather automatically construct a dataset of
image patches triplets. Each triplet consists of an an-
chor patch, one positive patch (which is a perspective
transformation of the anchor) and one negative patch.
During the training process, the network parameters
are trained with an adaptive triplet loss and updated
so that for a given training triplet, the anchor-positive
distance is minimized, while the anchor-negative dis-
tance is maximized. The learned CNN features are
then used to classify the dataset and estimate the
inter-class separability and intra-class compactness.
(128,128,1)
(124,124,16)
conv (5x5)
ReLU
MaxPool
(2x2)
(62,62,16)
(1,1,2048)
FCL 1
ReLU
(1,1,1024)
(1,1,512)
(1,1,128)
Output
Input
FCL 2
ReLU
FCL 3
ReLU
FCL 4
ReLU
Figure 1: Proposed CNN architecture. For a given 128 ×
128 patch, a 128-dimensional feature vector is provided in
the embedding space.
A comparison with the classical SIFT descriptor is
carried out in terms of precision/recall as in (Miko-
lajczyk and Schmid, 2005) to evaluate the efficiency
of the learned CNN features.
The organization of this paper is as follows: Sec-
tion 2 describes the proposed approach. Experimen-
tal results are presented and discussed in section 3.
Finally, section 4 provides conclusions and future
works.
2 PROPOSED APPROACH
This section details the proposed CNN architecture
as well as training steps that lead to a discriminative
feature space to describe all extracted patches around
image interest points. The main idea consists then in
training the proposed CNN with a set of patch triplets
in order to optimize an adaptive triplet loss that en-
courages similar patches to be closer in the feature
space. Comes after the feature matching step which is
achieved via computation of Euclidean distances be-
tween features in the embedding space.
2.1 CNN Architecture
The CNN proposed in this work is applied on input
patches of size 128 × 128. Figure 1 details the archi-
tecture of the proposed CNN. First, a 2D convolution
followed by a ReLU activation layer is applied to the
input data. Comes after a (2 × 2) max-pooling. Af-
ter that, 4 Fully Connected Layers (FCLs) intercepted
with ReLU activation layers are used. Finally, a nor-
malization of the data has been carried on. This archi-
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
926
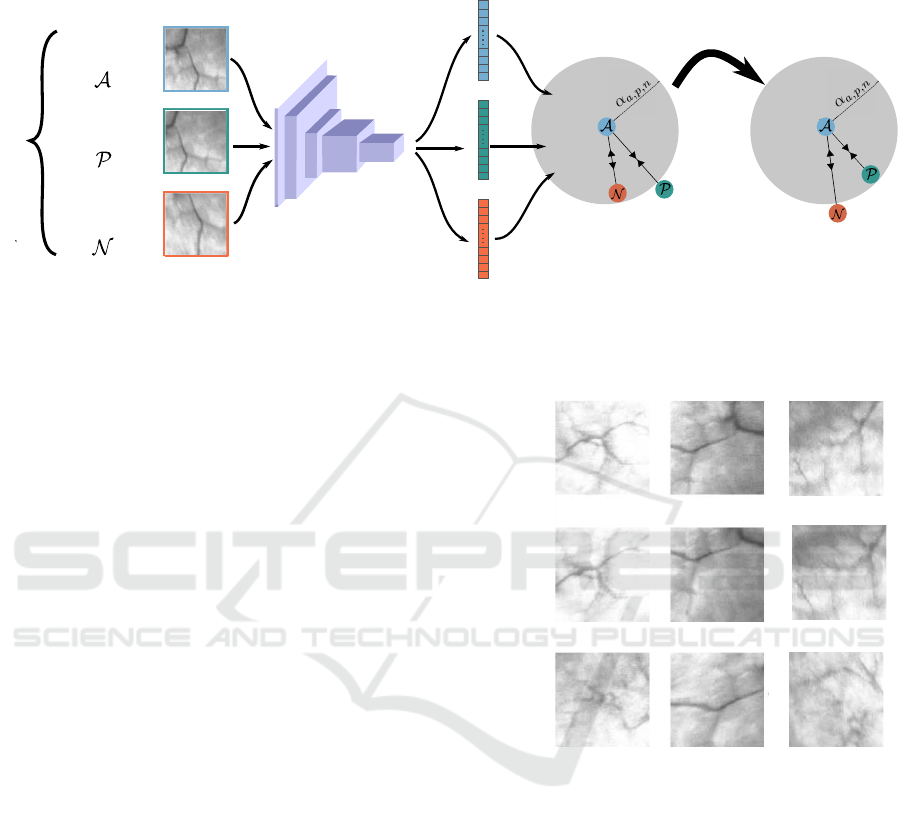
triplet loss
embedding
space
CNN
triplet patches
anchor
positive
negative
TRAINING
triplet loss
Figure 2: Triplet loss training approach. Features extracted from anchor, positive and negative samples are fed to the triplet
loss and trained so that the anchor-positive distance is minimized and the anchor-negative one is maximized in the feature
space. A more discriminative embedding space is thus obtained.
tecture provides at the end a 128- dimensional feature
vector.
2.2 CNN Training
The generation of the training dataset is explained as
follows: First, the raw data is converted from color
to gray-scale images. The contrast of the images is
then enhanced. After that, image interest points are
detected without loss of generality via SIFT detec-
tor. Anchor patches are gathered by cropping 128 by
128 pixels around the detected image interest points.
It is noticeable that organ surfaces are typically im-
aged from various angles and perspectives. In or-
der to get the corresponding positive patches, we ap-
ply pseudo randomly generated perspective transfor-
mations on the set of collected anchor patches. In
fact, such transformations could simulate well typi-
cal angles/perspective changes in endoscopic videos.
The obtained positive patches have the advantage to
appear very similar to anchor patches. The nega-
tive ones, on the other hand, are chosen randomly
from the rest of the anchors and positive samples of
other triplets. Figure 3 shows some examples of con-
structed patch triplets.
Depending on the desired prediction task, many
loss functions have been proposed in the literature.
In this work, we opted for the triplet loss to train
our model. Triplet loss, introduced in (Schroff et al.,
2015) as FaceNet model, has been successfully used
in several tasks (Vygon and Mikhaylovskiy, 2021;
Kumar et al., 2021). It has the advantage to enforce
the inter-class separability as well as the intra-class
compactness. The main idea consists in comparing
a baseline (anchor) input to a positive (truthy) input
and a negative (falsy) input. During the optimization
Sample 1 Sample 2
Anchor
Positive
Negative
Sample 3
Figure 3: Three samples (i.e., sample 1, 2 and 3) of col-
lected triplet patches. An anchor is a patch cropped around
an image interest point, already detected without loss of
generality via SIFT detector. While the positive sample is
a perspective transformation of the respective anchor, the
negative one is chosen randomly from the rest of the an-
chors and positive samples of another triplet.
process, the network parameters are updated so that
the anchor-positive input distance is minimized, while
the anchor-negative input distance is maximized. Fig-
ure 2 illustrates the complete principle of the adaptive
triplet loss.
The triplet loss proposed in this work is defined by
the following equation:
L =
∑
a,p,n∈{A,P,N}
(k f (a) − f (p)k
2
2
− k f (a) − f (n)k
2
2
+ α
a,p
)
(1)
Deep Features Extraction for Endoscopic Image Matching
927

where {A, P, N} denotes the triplets set: A, P and N
are respectively the anchor, positive and negative in-
puts. k.k
2
is the Euclidean distance. The function f (.)
stands for the feature embedding function that maps a
patch in the Euclidean embedding space via the CNN
architecture illustrated in Figure 1. Minimizing L en-
forces the maximization of the Euclidean distance be-
tween patches from different classes (anchor and neg-
ative) which should be greater than the distance be-
tween anchor and positive features in the embedding
space. In other words, the optimization process aims
at updating the different network parameters so that
the patches A and P become closer while A and N are
further apart in the Euclidean embedding space.
The term α
a,p
in Eq.(1) refers to a dynamic mar-
gin. It is designed as follows:
α
a,p
=
k f (a) − f (p)k
2
2
2
. (2)
In this work, we have noticed experimentally that the
use of adaptive margin is more advantageous than the
fixed ones.
It is worth to note that adaptive margins have been
used in many applications such as person reidentifi-
cation (Li et al., 2018), image retrieval (Zhao et al.,
2019). Promising results were obtained showing that
adaptive triplet loss encourages the learned image em-
bedding models to generalize well on cross-domain
data.
2.3 Feature Matching
The step after feature extraction with the trained CNN
is feature matching where the Euclidean distance be-
tween patches P
t
and P
t+1
cropped from two consec-
utive frames is computed in the embedding space ac-
cording to the following equation:
D( f (P
t+1
), f (P
t
)) =
k
f (P
t+1
) − f (P
t
)
k
2
2
(3)
This definition transforms finding patch matching
problem into a nearest neighbor search problem in the
Euclidean space. In this work, we utilized Nearest
Neighbor with a Threshold (NNT) to match patches
in which matching pairs is identified if the distance is
smaller than a certain threshold
1
. Note that, thresh-
olding based matching is commonly used to evaluate
descriptor performances (Lee and Park, 2014).
1
In our case, we set experimentally the NNT threshold
to 5.
3 EXPERIMENTAL RESULTS
AND DISCUSSIONS
We ran a series of experiments to evaluate the per-
formance of the proposed endoscopic image match-
ing in terms of True/False image interest point match-
ing. We additionally compared the obtained results to
SIFT descriptor, one of the well known state-of-the-
art hand-crafted image descriptors.
3.1 Dataset Construction
To evaluate the proposed model, we collected a set
of 300 image interest points from clinical patient en-
doscopic data with very different appearances. After
that, positive and negative patches are constructed as
already explained in 2. Figure 4 illustrates the dataset
construction process.
3.2 CNN Training
When applying the CNN architecture proposed in this
paper to the dataset described previously, the loss dis-
played in Figure 5 has been obtained, showing the
convergence of the model. The data has been trained
with a batch size of 36 using Stochastic Gradient De-
scent with an initial learning rate of 0.001 and a mo-
mentum set of 0.9. To avoid the over fitting of the
model to the selected patch triplets, we update the
training dataset each 50 epochs. The total number of
epochs is 250. The total number of triplets is over
15000 triplets.
3.3 Obtained Results
The proposed model is evaluated in terms of:
• Discriminative ability of the obtained embedding
space or in other words the representativeness of
the generated features.
• Robustness to typical angles/perspective changes
in endoscopic videos.
3.3.1 Discriminative Ability of the Embedding
Space
In order to measure the ability of the proposed method
to generate a discriminative embedding space rep-
resentation of the dataset, we define three types of
triplets; hard, semi-hard, and easy samples. The de-
gree of easiness/ hardness is expressed in terms of
inter-class separability and intra-class compactness.
More Details about the different sample types are
given bellow:
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
928
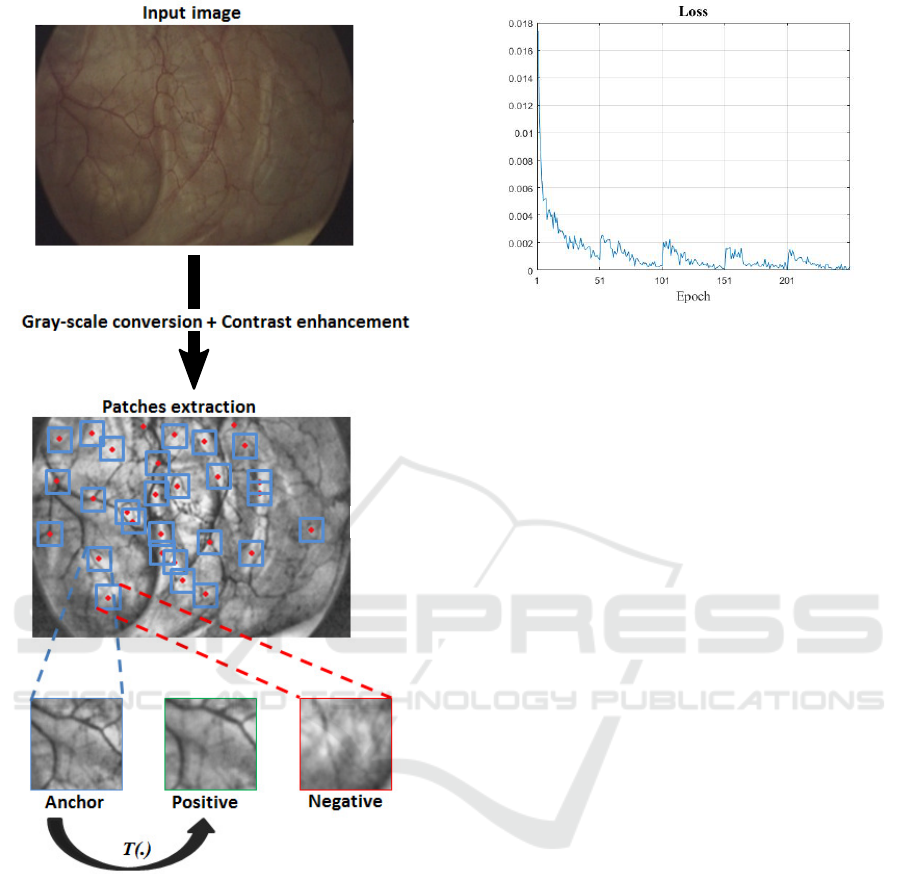
Figure 4: After conversion to gray-scale level and contrast
enhancement, image interest points are detected. Anchors
are obtained by cropping 128 by 128 pixels around the de-
tected image interest points. While positive patches are per-
spective transformation of the corresponding anchors (via
T (.)), the negative ones are selected randomly from the rest
of anchors and positives.
• Hard Samples: in this case, the samples will
be matched as false positives since the anchor-
positive distance is greater than the anchor-
negative one.
k f (a) − f (p)k
2
2
> k f (a) − f (n)k
2
2
• Semi-hard Samples: in this case, the anchor-
positive distance is still smaller than the anchor-
negative one but becomes larger when adding the
Figure 5: Convergence of the loss of the proposed approach.
An update of the training dataset is carried on every 50
epochs to avoid the over fitting. We notice that the loss
increases systematically on each dataset update but starts
again to decrease.
adaptive margin. Formally:
k f (a) − f (p)k
2
2
+ α
a,p
> k f (a) − f (n)k
2
2
.
• Easy Samples: these samples should also be
called safe samples. In here, it is easy to sepa-
rate the negative patch. In face, these samples are
characterized by the fact that the anchor-positive
distance remains always smaller than the anchor-
negative one even when adding the adaptive mar-
gin. Mathematically speaking:
k f (a) − f (p)k
2
2
+ α
a,p
< k f (a) − f (n)k
2
2
.
From the definitions given above, it is clear that the
larger the proportion of easy samples is, the more dis-
criminative the embedding space is, and the respec-
tively the larger the proportion of the hard samples is,
the less discriminative the embedding space is.
During the training process we monitor the pro-
portion (or percentage) of each of the three triplet
types. The idea is to check if during the training the
obtained feature space allows to increase the propor-
tion of easy samples at the expense of semi-hard and
hard samples, resulting thus in a more discriminative
feature space. Figure 6 summarizes the updates of the
easy-to-hard partition of the triplet samples during the
training step.
As depicted in Figure 6, we could notice that
the semi-hard samples proportion decreases dramat-
ically, while the easy samples one gets higher, reach-
ing more than 90% at the end of the training process.
This means that through the optimization process, the
different network parameters are updated so that the
learned features transfer samples from the hard, semi-
hard packages to the easy one, where the matching be-
tween the anchor and positive patches becomes more
intuitive.
Deep Features Extraction for Endoscopic Image Matching
929
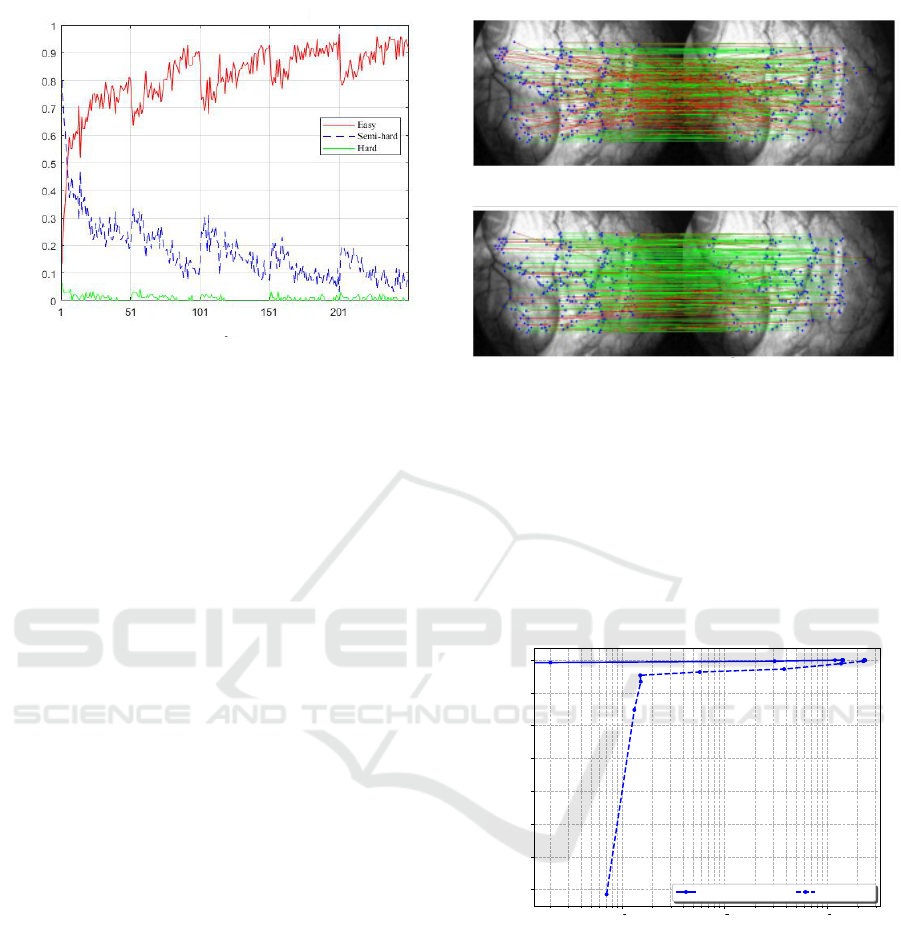
Percentage %
Epochs
Figure 6: Easy-to-hard partition of the triplet samples. We
observe that over training epochs a more discriminative
space representation is estimated allowing the an easy sep-
aration between positive and negative samples. The propor-
tion of easy samples increases (reaching more than 90%) at
the expense of semi-hard and hard samples (together less
than 10%).
3.3.2 Robustness of the Model to Viewpoint
Changes
When using an endoscope, organ surfaces are typi-
cally imaged from various angles and perspectives,
resulting in a stream of video endoscopy. There-
fore, we evaluate the performance of our approach in
matching interest points in the same endoscopic video
in terms of robustness against viewpoint changes. We
also compare it to the well-known standard hand-
crafted descriptor SIFT.
To simulate typical viewpoint changes in endo-
scopic videos, we first detect image interest points and
then estimate homographies from real clinical data
between consecutive frames based on RANSAC al-
gorithm (Fischler and Bolles, 1981).
Ten sequences of human bladder endoscopic
videos from different patients have been used for this
purpose. Each sequence consists of thousands of
frames with a wide range of texture, luminosity, and
appearance. Homologous points matching between
consecutive frames has been manually performed to
collect the matching ground truth. Once homogra-
phies have been estimated, we randomly apply them
to image sequences. For each pair of images ( i.e.,
original and warped images), image interest points are
detected. Then, point matching is performed twice,
using our method and SIFT method. Evaluation is
carried out qualitatively and quantitatively to compare
the two methods.
Figure 7 depicts the qualitative evaluation of the
SIFT descriptor and the proposed one when match-
ing two endoscopic frames. Green and red lines re-
(a) SIFT matching.
(b) Our matching.
Figure 7: Qualitative evaluation of the matching perfor-
mances carried out between two endoscopic frames. Im-
age interest points are colored in blue. Green and red lines
refer respectively to correct and wrong matches. The best
matching quality is reached with our approach.
fer respectively to correct and wrong matches. Com-
pared to SIFT, we can observe that a larger number
of correct matches (green lines) is achieved with our
approach.
10
3
10
2
10
1
1-Precision
0.6 5
0.7 0
0.7 5
0.8 0
0.8 5
0.9 0
0.9 5
1.0 0
Recall
Precision-Recall Curv es w it h NNT
Prop osed SIFT
Figure 8: Quantitative evaluation: Recall/precision com-
puted for SIFT and proposed approach when varying view-
point angles. The proposed descriptor outperforms SIFT,
reaching the maximal recall value for a precision value of
0.97.
For quantitative evaluation, we compare our
learned features matching performance to the SIFT
one in terms of recall with regard to precision. The
obtained results are depicted in Figure 8. We can
see that our method outperforms SIFT reaching the
top recall value for a precision value of 0.97. An-
other advantage of the method over hand-crafted ap-
proaches such as SIFT is that the proposed training
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
930

scheme does not require manual annotation, allowing
for additional few steps of fine tuning without manual
intervention for a better adaptation of the CNN model
for the sequence specificity without loss of generality
of the approach.
4 CONCLUSIONS AND FUTURE
WORKS
In this work, we propose the first deep learning
based approach for endoscopic image matching. For
this purpose, a triplet-based dataset of image patches
triplet has been firstly constructed. After that, to train
the CNN, an adaptive triplet loss has been designed to
improve the inter-class separability as well as the inter
class compactness leading to a discriminative feature
space. We assessed the robustness of the proposed ap-
proach against viewpoint changes and compared the
obtained performances to SIFT, one of the most suc-
cessful state-of-art descriptors.
Our further work will be focused on exploring
graph neural networks to integrate neighboring image
interest points in order to improve the discriminative
ability of the model. We intend also to test our ap-
proach on other endoscopic data captured for different
organs.
REFERENCES
Ait-Aoudia, S., Mahiou, R., Djebli, H., and Guerrout, E.-
H. (2012). Satellite and Aerial Image Mosaicing - A
Comparative Insight. In 2012 16th International Con-
ference on Information Visualisation, pages 652–657,
Montpellier, France. IEEE.
Ali, S., Zhou, F., Bailey, A., Braden, B., East, J. E., Lu,
X., and Rittscher, J. (2021). A deep learning frame-
work for quality assessment and restoration in video
endoscopy. Medical Image Analysis, 68:101900. Pub-
lisher: Elsevier.
Bay, H., Ess, A., Tuytelaars, T., and Van Gool, L. (2008).
Speeded-Up Robust Features (SURF). Computer Vi-
sion and Image Understanding, 110(3):346–359.
Behrens, A., Bommes, M., Stehle, T., Gross, S., Leonhardt,
S., and Aach, T. (2010). Real-time image composition
of bladder mosaics in fluorescence endoscopy. Com-
puter Science - Research and Development.
Behrens, A., Stehle, T., Gross, S., and Aach, T. (2009).
Local and global panoramic imaging for fluorescence
bladder endoscopy. In 2009 Annual International
Conference of the IEEE Engineering in Medicine and
Biology Society, pages 6990–6993. IEEE.
Ben-Hamadou, A., Daul, C., and Soussen, C. (2016). Con-
struction of extended 3d field of views of the inter-
nal bladder wall surface: A proof of concept. 3D Re-
search, 7(3):1–23.
Bergen, T., Wittenberg, T., M
¨
unzenmayer, C., Chen, C.
C. G., and Hager, G. D. (2013). A graph-based ap-
proach for local and global panorama imaging in cys-
toscopy. In Medical Imaging 2013: Image-Guided
Procedures, Robotic Interventions, and Modeling,
volume 8671, page 86711K. International Society for
Optics and Photonics.
Daul, C., Blondel, W., Ben-Hamadou, A., Miranda-Luna,
R., Soussen, C., Wolf, D., and Guillemin, F. (2010).
From 2d towards 3d cartography of hollow organs. In
2010 7th International Conference on Electrical En-
gineering Computing Science and Automatic Control,
pages 285–293. IEEE.
Du, P., Zhou, Y., Xing, Q., and Hu, X. (2011). Improved
SIFT matching algorithm for 3D reconstruction from
endoscopic images. In Proceedings of the 10th In-
ternational Conference on Virtual Reality Continuum
and Its Applications in Industry, pages 561–564.
Elibol, A., Kim, J., Gracias, N., and Garcia, R. (2017). Fast
Underwater Image Mosaicing through Submapping.
Journal of Intelligent & Robotic Systems, 85(1):167–
187.
Fischler, M. A. and Bolles, R. C. (1981). Random sample
consensus: a paradigm for model fitting with appli-
cations to image analysis and automated cartography.
Communications of the ACM, 24(6):381–395. Pub-
lisher: ACM New York, NY, USA.
Ghosh, T., Li, L., and Chakareski, J. (2018). Effective
deep learning for semantic segmentation based bleed-
ing zone detection in capsule endoscopy images. In
2018 25th IEEE International Conference on Image
Processing (ICIP), pages 3034–3038. IEEE.
Hossein-Nejad, Z. and Nasri, M. (2018). A-RANSAC:
Adaptive random sample consensus method in mul-
timodal retinal image registration. Biomedical Signal
Processing and Control, 45:325–338. Publisher: El-
sevier.
Kim, Y. J., Bae, J. P., Chung, J.-W., Park, D. K., Kim, K. G.,
and Kim, Y. J. (2021). New polyp image classifica-
tion technique using transfer learning of network-in-
network structure in endoscopic images. Scientific Re-
ports, 11(1):1–8. Publisher: Nature Publishing Group.
Kumar, P., Jain, S., Raman, B., Roy, P. P., and Iwamura,
M. (2021). End-to-end Triplet Loss based Emotion
Embedding System for Speech Emotion Recognition.
In 2020 25th International Conference on Pattern
Recognition (ICPR), pages 8766–8773. IEEE.
Lai, S.-C., Kong, M., Lam, K.-M., and Li, D. (2019).
High-resolution face recognition via deep pore-feature
matching. In 2019 IEEE International Conference on
Image Processing (ICIP), pages 3477–3481. IEEE.
Lee, M. H. and Park, I. K. (2014). Performance evaluation
of local descriptors for affine invariant region detector.
In Asian Conference on Computer Vision, pages 630–
643. Springer.
Li, Z., Sang, N., Chen, K., Gao, C., and Wang, R. (2018).
Learning deep features with adaptive triplet loss for
person reidentification. In MIPPR 2017: Pattern
Deep Features Extraction for Endoscopic Image Matching
931

Recognition and Computer Vision, volume 10609,
page 106090G. International Society for Optics and
Photonics.
Liu, G., Hua, J., Wu, Z., Meng, T., Sun, M., Huang, P., He,
X., Sun, W., Li, X., and Chen, Y. (2020). Automatic
classification of esophageal lesions in endoscopic im-
ages using a convolutional neural network. Annals of
translational medicine, 8(7). Publisher: AME Publi-
cations.
Liu, Y., Xu, X., and Li, F. (2018). Image feature matching
based on deep learning. In 2018 IEEE 4th Interna-
tional Conference on Computer and Communications
(ICCC), pages 1752–1756. IEEE.
Lopez, F., Varela, A., Hinojosa, O., Mendez, M., Trinh, D.-
H., ElBeze, J., Hubert, J., Estrade, V., Gonzalez, M.,
Ochoa, G., and Daul, C. (2021). Assessing deep learn-
ing methods for the identification of kidney stones
in endoscopic images. arXiv:2103.01146 [cs, eess].
arXiv: 2103.01146.
Lowe, D. G. (2004). Distinctive image features from scale-
invariant keypoints. International journal of computer
vision, 60(2):91–110. Publisher: Springer.
Martinez, A., Trinh, D.-H., El Beze, J., Hubert, J., Es-
chwege, P., Estrade, V., Aguilar, L., Daul, C., and
Ochoa, G. (2020). Towards an automated classifi-
cation method for ureteroscopic kidney stone images
using ensemble learning. In 2020 42nd Annual In-
ternational Conference of the IEEE Engineering in
Medicine & Biology Society (EMBC), pages 1936–
1939, Montreal, QC, Canada. IEEE.
Mikolajczyk, K. and Schmid, C. (2005). A perfor-
mance evaluation of local descriptors. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
27(10):1615–1630. Conference Name: IEEE Trans-
actions on Pattern Analysis and Machine Intelligence.
Schroff, F., Kalenichenko, D., and Philbin, J. (2015).
Facenet: A unified embedding for face recognition
and clustering. In Proceedings of the IEEE conference
on computer vision and pattern recognition, pages
815–823.
Vygon, R. and Mikhaylovskiy, N. (2021). Learning Ef-
ficient Representations for Keyword Spotting with
Triplet Loss. arXiv preprint arXiv:2101.04792.
Weibel, T., Daul, C., Wolf, D., R
¨
osch, R., and Ben-
Hamadou, A. (2010). Endoscopic bladder image reg-
istration using sparse graph cuts. In 2010 IEEE Inter-
national Conference on Image Processing, pages 157–
160. IEEE.
Zhao, X., Qi, H., Luo, R., and Davis, L. (2019). A weakly
supervised adaptive triplet loss for deep metric learn-
ing. In Proceedings of the IEEE/CVF International
Conference on Computer Vision Workshops, pages 0–
0.
Zou, S., Long, M., Wang, X., Xie, X., Li, G., and Wang, Z.
(2019). A CNN-Based Blind Denoising Method for
Endoscopic Images. In 2019 IEEE Biomedical Cir-
cuits and Systems Conference (BioCAS), pages 1–4.
IEEE.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
932
