
Simulating the Doctor’s Behaviour: A Preliminary Study on the
Identification of Atrial Fibrillation through Combined Analysis of Heart
Rate and Beat Morphology
Gennaro Laudato
1 a
, Giovanni Rosa
1 b
, Giovanni Capobianco
1
, Angela Rita Colavita
2
,
Arianna Dal Forno
1
c
, Fabio Divino
1 d
, Claudio Lupi
1 e
, Remo Pareschi
1 f
, Stefano Ricciardi
1
,
Luca Romagnoli
1
, Simone Scalabrino
1 g
, Cecilia Tomassini
1 h
and Rocco Oliveto
1 i
1
STAKE Lab, University of Molise, Pesche (IS), Italy
2
ASREM – Regione Molise, Italy
Keywords:
Recommender System, Deep Learning, ECG Analysis, Atrial Fibrillation, Arrhythmia.
Abstract:
Atrial fibrillation (AF) is a medical disorder that affects the atria of the heart. AF has emerged as a world-
wide cardiovascular epidemic affecting more than 33 million people around the world. Several automated
approaches based on the analysis of the ECG have been proposed to facilitate the manual identification of AF
episodes. Especially, such approaches analyze the heartbeat morphology (absence of P-wave) or the heart rate
(presence of arrhythmia). In this article, we present AMELIA (AutoMatic dEtection of atriaL fIbrillation for
heAlthcare), an approach that simulates the doctor’s behavior by considering both the sources of information
in a combined way. AMELIA is basically composed of two components; one integrating a LSTM (Long
Short-Term Memory) Recurrent Neural Network (RNN) and the second integrating a rhythm analyzer. When
the RNN reveals a heartbeat with abnormal morphology, the rhythm analyzer is activated to verify whether
or not there is a simultaneous arrhythmia. AMELIA has been experimented by using well-known datasets,
namely Physionet-AF and NSR-DB. The achieved results provide evidence of the potential benefits of the ap-
proach, especially regarding sensitivity. AMELIA has an incredibly high potential to be used in applications
of continuous monitoring, where the detection of AF episodes is a fundamental and crucial activity.
1 INTRODUCTION
In modern healthcare systems the vital signals of pa-
tients are acquired, collected, and analyzed within the
system itself.
This is the case of ATTICUS (Laudato et al.,
2021), an innovative tele-service and remote monitor-
ing system for ambient-assisted living based on the
analysis of vital and behavioral parameters. The data
a
https://orcid.org/0000-0002-3776-2848
b
https://orcid.org/0000-0002-5241-1608
c
https://orcid.org/0000-0003-0500-3852
d
https://orcid.org/0000-0003-4107-3727
e
https://orcid.org/0000-0001-5166-1130
f
https://orcid.org/0000-0002-4912-582x
g
https://orcid.org/0000-0003-1764-9685
h
https://orcid.org/0000-0002-2819-7779
i
https://orcid.org/0000-0002-7995-8582
are acquired through a smart t-shirt (Balestrieri et al.,
2019; De Vito et al., 2021) and then transmitted to an
Ambient Intelligence device located nearby, which,
in turn, predicts potentially anomalous situations and
it communicates them to a Decision Support System
(DSS). Such a system can perform deeper and more
accurate analysis and, if it confirms the anomaly, it
can alert a monitoring station in which human experts
(e.g., doctors) manually analyze the data and plan an
intervention.
In this paper, we present an approach that we aim
at integrating into the DSS of ATTICUS. The ap-
proach is in charge of analyzing the ECG to iden-
tify atrial fibrillation (AF) events, an abnormal heart
rhythm characterized by rapid and irregular beating
of the atria. The process of AF episodes diagno-
sis involves two ECG sources of information: (i)
morphology-based, because during an AF episode,
446
Laudato, G., Rosa, G., Capobianco, G., Colavita, A., Forno, A., Divino, F., Lupi, C., Pareschi, R., Ricciardi, S., Romagnoli, L., Scalabrino, S., Tomassini, C. and Oliveto, R.
Simulating the Doctor’s Behaviour: A Preliminary Study on the Identification of Atrial Fibrillation through Combined Analysis of Heart Rate and Beat Morphology.
DOI: 10.5220/0010823900003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 446-453
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

fluctuating waveforms instead of P waves can be ob-
served and (ii) rhythm-based, because of the heart rate
irregularity, which may appear.
A lot of effort was devoted by the research com-
munity to the definition of approaches for the auto-
matic detection of AF by using Machine Learning
(ML) techniques based on one of the above sources
of information. Indeed, a widespread approach is
the Support Vector Machines (SVM) (Sepulveda-
Suescun et al., 2017; Islam et al., 2017; Padma-
vathi and Ramakrishna, 2015), while other authors
chose Neural Network (NN) to classify ECG seg-
ments (Yuan et al., 2016; Xiong et al., 2017) or novel
recursive algorithms (Zhou et al., 2015). The most
common features for the ML tools used in these
methods are based on RR Intervals (RRI) analysis.
There are also approaches, such as the one by Padma-
vathia and Ramakrishnab (Padmavathi and Ramakr-
ishna, 2015), where it is proposed the use of autore-
gressive coefficients as derived morphological ECG
features.
Despite the high accuracy of the above methods,
we conjecture that there is still room for improve-
ment. We believe that an approach – based on the
combination of morphological and rhythmic analysis
– can reproduce the exact procedure used by cardiol-
ogists when manually checking an ECG for AF diag-
nosis. Our conjecture is supported by the results of
previous works proposed in Laudato et al. (2020b,a),
where a machine learning approach, named MOR-
PHYTHM was defined to combine morphological and
rhythmic information to support the identification of
AF events. Despite the promising results achieved,
the main limitation of such an approach was repre-
sented by the difficulties to explain a specific predic-
tion. Indeed, all the features extracted from the ECG
were put together in one single learning algorithm.
This made it difficult to identify the event that trig-
gered the prediction.
Based on the willingness to have an accurate
and explainable method for detecting AF events, we
present AMELIA (AutoMatic dEtection of atriaL
fIbrillation for heAlthcare). AMELIA is an auto-
mated AF detector based on (i) an LSTM Recurrent
Neural Network (Hochreiter and Schmidhuber, 1997)
for the ECG Morphology classification and on (ii)
statistical heuristics to identify arrhythmia. The pro-
posed approach aims at simulating as much as pos-
sible the doctor’s behaviour during the detection of
AF episodes. Especially, AMELIA first analyzes the
morphology of the heartbeat (as it is and not in terms
of derived features as shown in the literature) in order
to identify the absence of p-wave and then confirm the
anomaly by checking the presence of arrhythmia.
The proposed approach was experimented on
two publicly accessible sets of clinical data (MIT-
BIH Atrial Fibrillation Database
1
and MIT-BIH Nor-
mal Sinus Rhythm Database
2
). The accuracy of
AMELIA was compared with the work by Zhou
et al. (2015), one of the most accurate methods of
AF detection based on RRI analysis (therefore based
on only rhythmic features) and with the work re-
cently proposed by Laudato et al. (2020b) which, as
AMELIA, also takes in consideration morphological
and rhythmic features. The results reported in Sec-
tion 4 show that it is possible to achieve benefits with
respect to the chosen baselines.
We believe that AMELIA can be better em-
ployed in telemedicine applications, where e-AI
(explainable-Artificial Intelligence) is often a strong
requirement. Indeed, AMELIA– thanks to the con-
ceptual and de facto separation of the data sources
between rhythmic and morphological – can provide
highly accurate information in the process of diag-
nosis. For example, AMELIA– beyond the genera-
tion of a warning indicating a potential AF episode –
can provide the additional information of which is the
heartbeats not showing a P wave with a high accuracy.
The premise that AMELIA can reproduce the ex-
act procedure used by cardiologists is based on the
consideration that the approach takes as input a se-
quence of heartbeats that are submitted only to noise-
removal and downsampling processing stages, there-
fore preserving the ECG shape. AMELIA aims
at simulate the manual diagnosis by observing the
rhythm and the shape of a pattern of heartbeats.
The rest of the paper is structured as follows: Sec-
tion 2 presents the proposed approach for AF detec-
tion, Section 3 describes the experimental choices for
the design of the study, while Section 4 presents the
results of the evaluation of the proposed approach on
the Physionet data set. Finally, Section 6 concludes
the paper by discussing the results and by reporting
all the potential future works which can be undertaken
with AMELIA in the context of ATTICUS (Balestri-
eri et al., 2019; Laudato et al., 2021).
2 AMELIA
An AF episode is diagnosed by a doctor when the
morphology of the heartbeat is abnormal (no P-wave),
RR intervals are irregularly irregular, and f-waves ap-
pear. AMELIA aims at simulating as much as possi-
ble such a behavior. The workflow of AMELIA is
1
https://physionet.org/physiobank/database/afdb/
2
https://physionet.org/physiobank/database/nsrdb/
Simulating the Doctor’s Behaviour: A Preliminary Study on the Identification of Atrial Fibrillation through Combined Analysis of Heart
Rate and Beat Morphology
447
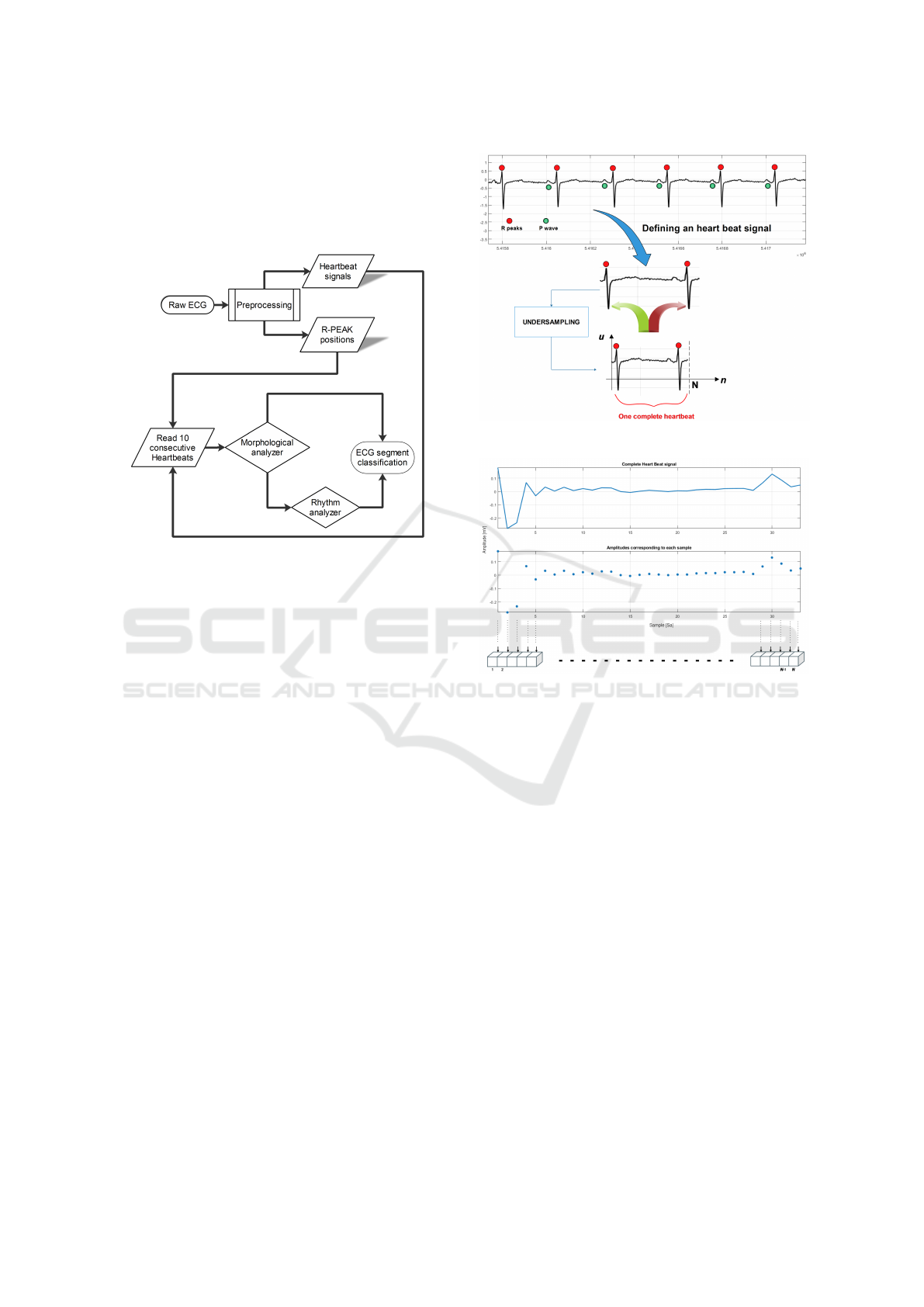
depicted in Fig. 1. In the preprocessing stage,
AMELIA extracts all the heartbeat signals and all the
R peak positions from a raw single lead digital ECG
acquired with a given sampling frequency. These sig-
nals are submitted to a Morphology analyzer.
Figure 1: AMELIA workflow.
If the morphology of the heartbeat is abnormal,
the Morphology analyzer triggers the Rhythm ana-
lyzer. The Rhythm analyzer takes as an input the
extracted R peak positions and tries to consolidate
the initial warning identified by the Morphology an-
alyzer. If the Rhythm analyzer identifies through
the analysis of ten consecutive R-R intervals an ar-
rhythmia, then an AF episode is identified. Otherwise,
the initial warning of the Morphology analyzer is re-
jected. In this case, the abnormal morphology of the
heartbeat could be due to the wrong classification of
the NN or just to some noise in the ECG.
2.1 Definition of a Heartbeat
It is necessary to clarify the concept of heart beat sig-
nal. In AMELIA, a heartbeat signal is a raw ECG
segment included between two successive R peaks
(see Fig. 2). The choice to define a heartbeat signal
in this way is due to the consideration that the mor-
phological features – observable during AF episodes
– are (i) the absence of P wave and (ii) the poten-
tial fibrillation waves in its place. The concept of the
heartbeat signal is also faced in the work by Xu et
al. (Xu et al., 2018) with the difference that the au-
thors define it as the signal between the two middle
points of three consecutive R peaks. We decided to
work with heart dynamics included between two suc-
cessive R peaks because the fibrillating phenomena
are inscribed between those waves. We used the Pan-
Figure 2: Definition of heartbeat signal in AMELIA.
Figure 3: Representation of a complete heartbeat in
AMELIA.
Tompkins method (Pan and Tompkins, 1985) to ob-
tain all the expected heartbeat signals of a given full
ECG signal. We opted for the validation of AMELIA
in the online scenario (worst case), therefore without
using the beat annotations available from Physionet.
A complete heartbeat is represented by a vector
defined as:
hbs = [u
1
, u
2
, ..., u
N
] (1)
where u
1
and u
N
are the raw amplitudes of the
samples corresponding to the position of the left and
right R peak, respectively (see Fig. 3).
In order to provide fixed-length instances to the
Morphology analyzer component – all the heartbeat
signals were submitted to a process of down-sampling
(in section 4.3, more details are provided). N is the
fixed length of each complete heartbeat signal.
2.2 Morphology Analyser
The Morphology Analyser is in charge of analyzing
the morphology of a heartbeat. The input of this com-
ponent is represented by a heartbeat. Ideally, the out-
HEALTHINF 2022 - 15th International Conference on Health Informatics
448

put is no-AF if the morphology of the heartbeat is
normal and AF if the morphology of the heartbeat
does not have the P wave and shows fluctuating wave-
forms (f-waves), i.e., the morphological characteris-
tics of a heart beat in the presence of an AF event. The
morphology classification of the heartbeat is based on
a Recurrent Neural Network (RNN) (Hochreiter and
Schmidhuber, 1997) with multiple LSTM cells. The
choice is justified by the consideration that LSTM
cells better adapt to time-series classification (Karim
et al., 2017) (as in the case of ECG).
2.3 Rhythm Analyser
The Rhythm Analyser aims at identifying normal
rhythm or arrhythmia by evaluating a buffer of ten
successive heartbeats. It is worth noting that– based
on a consolidated opinion from cardiologists – ten
consecutive heartbeats can be can be deemed enough
to diagnose atrial fibrillation. This number is also
confirmed by the works in (Kurzweil et al., 2009;
Zurro et al., 1995) where a minimum of 3 and 6 suc-
cessive heartbeats was evaluated.
In details, the Rhythm Analyser classifies each
beat as short, long, or normal. Considering that the
normal heart rate during rest for teenagers is around
70-120 beat per minute (bpm) and adults is around
60-90 bpm (D. Limmer, 2005), each beat is classi-
fied as follows: short if bpm > 120, long if bpm <
50 and normal otherwise. Once the Rhythm Analyser
has buffered and labeled ten consecutive heartbeats,
computes the entropy of the buffer B.
2.4 Putting All Together
Algorithm 1 shows how the Morphology Analyser
and the Rhythm Analyser are combined in order to de-
tect AF events.
For each heartbeat signal, a fixed-length buffer
hbs
i
is instantiated, containing the amplitudes of the
signal. The buffer hbs
i
then is submitted to the Mor-
phology Analyser, which provides its classification.
When the morphology is classified as AF, a new
buffer of heartbeats is created. Once the buffer of
heartbeats has reached the max size (set as 10, in our
case), it is submitted to the Rhythm Analyser. Based
on the entropy information evaluated on the buffer, a
classification in terms of rhythm is provided. If also
the rhythm is identified as IRREGULAR, a warning is
generated.
Algorithm 1: Detection of Arrhythmia.
Require: ECG Raw ECG
HBS = ExtractHeartBeatSignals(ECG)
RRI = ExtractRRInterval(ECG)
for each hbs
i
∈ HBS do
Morphology = MorphologyAnalyser(hbs
i
)
if Morphology == AF then
buffer
i
←
/
0 new buffer for the i
th
heart
beat
BUFFERS ← BUFFERS ∪ buffer
i
end if
for each buffer
j
∈ BUFFERS do
buffer
j
← buffer
j
∪ RRI
i
if size(buffer
j
) == MAX SIZE then
Rhythm = RhythmAnalyser(buffer
j
)
if Rhythm == ABNORMAL then
GenerateWarning()
end if
BUFFERS ← BUFFERS \ buffer
j
end if
end for
end for
3 STUDY DESIGN
We compared AMELIA to the method proposed in
(Zhou et al., 2015), where AF episodes are identified
by using only an RRI analysis. Thus, in the context of
the study, we formulated the following research ques-
tion:
Does AMELIA outperform state-of-the-art
AF detection approaches?
We chose as baseline the approach by Zhou (2015)
et al. (Zhou et al., 2015) because in the state of the art,
it is one of the most accurate approaches based on RRI
analysis. We also keep a recent tool MORPHYTHM
(Laudato et al., 2020b) as a reference, because it is
based on a combination of rhythmic and morphologi-
cal analysis, too.
3.1 Context of the Study
The proposed approach was experimented on the
MIT-BIH AFDB (Goldberger et al., 2000). For
this DB, Physionet offers 25 2-lead ECG recordings.
These were acquired with a sampling frequency of
250 Hz, 12-bit resolution over a range of ± 10 mil-
livolts. In this preliminary study, we performed a de-
tection based on a single-lead ECG, thus we took into
account only the first lead. Furthermore, for this data
set, Physionet does not provide distinction among
beat types (but only in terms of rhythm); indeed, all
Simulating the Doctor’s Behaviour: A Preliminary Study on the Identification of Atrial Fibrillation through Combined Analysis of Heart
Rate and Beat Morphology
449
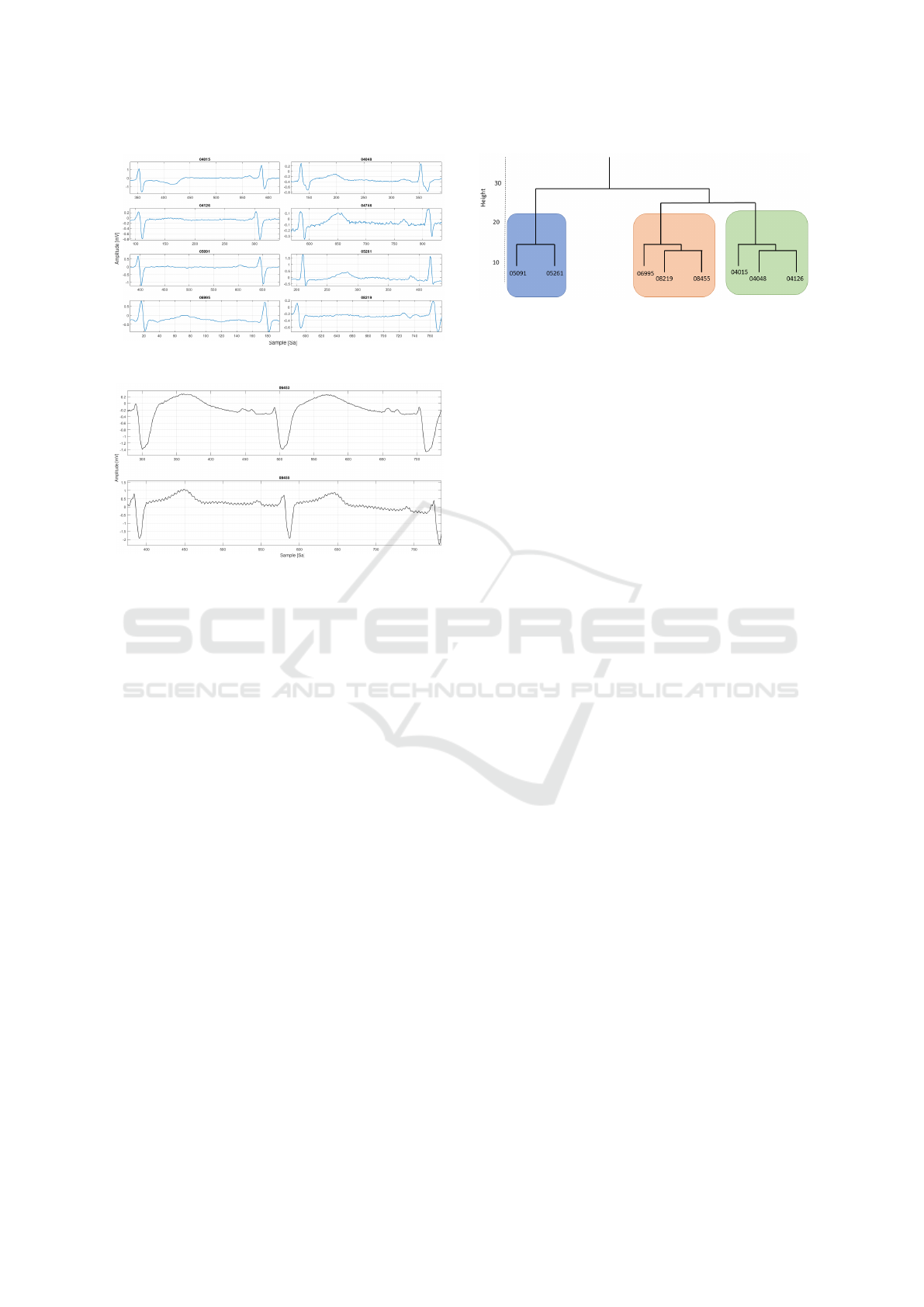
Figure 4: The records chosen for this study.
Figure 5: The records ignored for the study.
beats are labeled as normal. Considering the diver-
sity of the ECG shapes – in order to guarantee con-
sistency of information – we manually selected only
those recordings with a common shape. This choice is
due to specificity of AMELIA’s morphology compo-
nent. Thus, in this preliminary study, we considered
records #04015, #04048, #04126, #05091, #05261,
#06995, #08219, #08455. Indeed, this group of pa-
tients presents a high similarity between the ECG
waveform shapes (see Fig. 4). For the moment, the
other records were ignored. Examples of ignored
records are #06453 and #08455, where in the avail-
able signal 1 from the database, the shapes differ from
the ones of the above group of recordings (Fig. 5).
Each heartbeat signal was manually observed and
analyzed, with the help of a medical equipe. Only sig-
nals presenting a clear AF effect were selected. The
operation was carried out for all the chosen records.
A total of 1637 heartbeat signals from the 8 different
recordings were manually extracted. The minimum
length for each of these signals varies from 33 to 111
samples. By doing that, we obtained two types of sig-
nal: AF and Normal (no-AF) heart-beat signals.
To conclude the experiment, we tested the fi-
nal version of AMELIA also on the MIT-BIH Nor-
mal Sinus Rhythm Databases because it contains pa-
tients with a healthy ECG except for some no signif-
icant arrhythmia episodes. Therefore, at the end of
the study, we will experiment AMELIA under sev-
Figure 6: The dendrogram for the manually selected records
from AF Database, based on an AF heartbeat.
eral circumstances: the detection of AF and NO-AF
episodes. These latter include normal sinus rhythm
and pathological rhythm. Indeed, both the chosen
database include pathological rhythm different from
AF episodes. Specifically, the AFDB contains atrial
flutter episodes and the NSRDB arrhythmia episodes
(Goldberger et al., 2000).
3.2 Patient-centered Data Clustering
The 8 records were manually observed and selected.
Therefore, before reporting the classification results,
we aimed at validating the manual selections per-
formed in the previous steps. To do so, the data was
submitted to a clustering algorithm to assess effec-
tively if the 8 patients group together in an individ-
ual cluster. To this aim, we selected one AF-labelled
heartbeat from each of the chosen records, and we de-
scribed each of these heartbeats using several descrip-
tors, such as: entropy measures (e.g. the one proposed
in (Zhou et al., 2015)), Statistical Features (e.g. the
mean, variance, and norm of the amplitude samples),
Fast Fourier Transform and AR model coefficients.
After creating this data set, we applied a technique of
Hierarchical clustering with Euclidean distance as the
similarity function and the average as the agglomera-
tion method. We obtained the dendrogram depicted in
Fig. 6. Thus, even if they seemed to have a common
ECG waveform shape – when they are observed from
an AF heartbeat perspective – the clustering process
assigns them to distinct groups. This separation could
be due to physiological aspects that are embedded in
an ECG.
From this step of clustering, we obtained a refined
view of the groups to which our records belong. This
will be used to represent the results for each patient,
according to the reference cluster.
3.3 Training of the LSTM RNN
The training of the LSTM RNN was performed on
a balanced data set composed of 1637 instances for
the AF Class and 1653 for the no-AF Class. To the
HEALTHINF 2022 - 15th International Conference on Health Informatics
450

Figure 7: Examples of the manually selected instances from
the Physionet AFDB.
aim of guaranteeing an alignment, all the instances
were downsampled to 33 data points. An example
of selected and downsampled instances is depicted in
Fig. 7. The LSTM parameters were experimentally
defined through a trial & error approach. In order
to validate the network, a classical Leave-one-person-
out (LOPO-CV) cross-validation was applied to the
data set. LOPO-CV means that one person at a time
is left out from the training set, so that the training
set contains no data specific to the individual who is
being tested (the classifier was not tuned with the test
data of that person).
4 ANALYSIS OF THE RESULTS
We experimented with the proposed approach on
two freely accessible data set, the Physionet MIT-
BIH AFBD and the Normal Sinus Rhythm Database
(NSRDB) (Goldberger et al., 2000). We considered
the following classification metrics: TP (beat labeled
as AF and classified as AF), FP (beat labeled as no-
AF and classified as AF), TN (beat labeled as no-AF
and classified as no-AF), and FN (beat labeled as AF
and classified as no-AF).
Unfortunately – for the chosen baseline (Zhou
et al., 2015) – the authors did not report classifica-
tions at patient-level. Therefore, we replicated the
method and obtained the desired level of classifica-
tion. The classification results are shown in Tables
1, 2, 3 according to the clusters previously obtained.
The difference in the number of heartbeats is due to
the nature of AMELIA. Our online tool embeds the
Pan-Tompkins algorithm as a peak detector (while the
methods of the state of the art use the peak annota-
tions provided from Physionet). Therefore, even if
highly accurate, the performances of AMELIA inte-
grate an additive error due to potential wrong classi-
fications of this algorithm. Therefore, the only way
to compare AMELIA—with respect to the chosen
baselines—is to evaluate the overall statistics as: Sen-
Table 1: AMELIA classification performance compared to
the chosen baseline on MIT-BIH AF-db cluster 1.
Cluster 1 Method TP TN FP FN
#04015
AMELIA 500 42088 2886 25
MORPHYTHM 491 40650 2836 27
(Zhou et al., 2015) 478 40707 2779 40
#04048
AMELIA 792 38967 165 90
MORPHYTHM 443 38982 145 363
(Zhou et al., 2015) 419 38990 137 387
#04126
AMELIA 3345 39581 960 51
MORPHYTHM 3154 38149 1424 132
(Zhou et al., 2015) 3082 38743 830 204
Method Sens Spec Acc
AMELIA 0,965 0,968 0,968
MORPHYTHM 0,887 0,964 0,961
(Zhou et al., 2015) 0,863 0,969 0,965
Table 2: AMELIA classification performance compared to
the chosen baseline on MIT-BIH AF-db cluster 2.
Cluster 2 Method TP TN FP FN
#05091
AMELIA 44 35470 986 98
MORPHYTHM 0 36640 4 133
(Zhou et al., 2015) 0 36644 0 133
#05261
AMELIA 881 43739 1953 51
MORPHYTHM 766 43015 1595 157
(Zhou et al., 2015) 655 44215 395 268
Method Sens Spec Acc
AMELIA 0,861 0, 964 0, 963
MORPHYTHM 0, 725 0, 980 0, 977
(Zhou et al., 2015) 0, 620 0,995 0,990
sitivity =
T P
T P+F N
, Specificity =
T N
T N+FP
and Accuracy =
T P+T N
T P+T N+FP+FN
.
From the achieved results, it is possible to observe
that for records belonging to: Cluster 1: AMELIA
outperforms both the baseline and MORPHYTHM
in terms of all the metrics of validation; Cluster
2: AMELIA provides significantly higher sensitiv-
ity with slightly lower specificity and accuracy; Clus-
ter 3: AMELIA presents a significant loss, mostly in
terms of sensitivity and accuracy.
From the LOPO-CV cross-validation, we chose
the best network in terms of accuracy on the test data
set. With this, we experimented with the proposed
approach also on the MIT-BIH Normal Sinus Rhythm
Table 3: AMELIA classification performance compared to
the chosen baseline on MIT-BIH AF-db cluster 3.
Cluster 3 Method TP TN FP FN
#06995
AMELIA 11215 27160 490 17784
MORPHYTHM 27240 25901 1767 280
(Zhou et al., 2015) 27072 25648 2020 448
#08219
AMELIA 7946 42595 4286 6207
MORPHYTHM 13420 40934 4203 735
(Zhou et al., 2015) 12627 42637 2500 1528
#08455
AMELIA 32705 15265 22 12470
MORPHYTHM 44111 15244 45 151
(Zhou et al., 2015) 44103 15250 39 159
Method Sens Spec Acc
AMELIA 0, 587 0,947 0, 768
MORPHYTHM 0,986 0, 932 0, 959
(Zhou et al., 2015) 0, 975 0,948 0,962
Simulating the Doctor’s Behaviour: A Preliminary Study on the Identification of Atrial Fibrillation through Combined Analysis of Heart
Rate and Beat Morphology
451

Table 4: AMELIA’s accuracy on MIT-BIH NSR-db.
Record Rhythm Analyser AMELIA
ID # FP # FP
16265 2 0
16272 0 0
16273 1 0
16420 1 0
16483 0 0
16539 13 0
16773 0 0
16786 0 0
16795 4 0
17052 29 0
17453 0 0
18177 5 0
18184 0 0
19088 10 1
19090 0 0
19093 0 0
19140 0 0
19830 16 14
Databases. In this DB, all the recordings present a
shape with high similarity with respect to the ones
used in this study. The individuals included in the
NSRDB were found to have no significant arrhyth-
mia. Tables 4 show the results achieved on this DB.
This experimentation represents a boundary valida-
tion for our proposed methods because the goal is to
avoid all the phenomena with arrhythmia (different
from AF) by using the useful information provided
by the morphological module of AMELIA.
In this validation of AMELIA, we expected that
the proposed tool do not get confused with (not signif-
icant) arrhythmia episodes affecting the patient from
this data set. As we can see from the tables - by using
our rhythm analyzer - an arrhythmia can be detected
as an Atrial Fibrillation episode. With the introduc-
tion of the morphological Analyzer in AMELIA we
reduced the chance of misclassifying several heart-
beats.
5 THREATS TO VALIDITY
One of the limitations of the present study is that the
evaluation is performed on a reduced number of se-
lected recordings. This choice was due to the consid-
eration that the Neural Network—used in this context
as a morphology analyzer —is strictly dependent on
several features related to an ECG recording, such as
the lead, physiological aspects (smoker, BMI, etc.),
and the instrumentation used to acquire the ECG.
Therefore, we opted for manually selecting the
available recordings from the AF database to be in-
volved in this preliminary study. Of course, in the
real-world application of arrhythmia detection, the
detectors are subject to a broad range of beat mor-
phologies, including patients with ectopic beats, dif-
ferent types of arrhythmia, recordings corrupted by
noise, and so on. The evaluation results of this study
are to be intended only for a limited number of ECGs,
such as the ones with common features as the ones
selected for the validation of AMELIA. We also de-
cided to validate the manual selection of ECG record-
ings in order to assess the similarity between them.
From a refined perspective offered by the dendro-
gram, we observed that the 8 recordings could be fur-
ther grouped into three sub-clusters. Thanks to this re-
sult, we could report the evaluation in terms of ’clus-
ters’ in order to highlight the specificity of AMELIA
in the detection of AF episodes
The comparison with state of the art should be
considered only illustrative because the validation
was performed with different procedures: (i) Zhou
et al. (2015) used the Physionet Long-Term AF
Database
3
to tune their entropy threshold and the
MIT-BIH Atrial Fibrillation Database to validate it,
(ii) in the validation of MORPHYTHM (Laudato et al.,
2020b) a LOPO-CV among all the patients from the
MIT-BIH Atrial Fibrillation Database was performed
while (iii) in AMELIA a LOPO-CV between only the
selected patients from the MIT-BIH Atrial Fibrillation
Database was executed.
6 CONCLUSION
We have presented AMELIA, an approach for au-
tomated detection of atrial fibrillation in the context
of real-time monitoring of vital parameters. The ap-
proach is based on the combined use of two different
sources of information that have paramount impor-
tance in the detection of AF events: (i) morpholog-
ical analysis of the heartbeats; and (ii) RRI analysis.
An empirical study conducted on two different well-
known public data sets has shown the potential of the
proposed approach compared with the state-of-the-art
methods based on (i) just RRI analysis and (ii) on both
sources of information.
Future works will be devoted to extending the ex-
perimentation of AMELIA on other datasets.
Also, we plan to further improve the accuracy of
AMELIA, by replacing the manual selection of ECG
recordings with a fully automated process.
3
https://archive.physionet.org/physiobank/database/ltafdb/
HEALTHINF 2022 - 15th International Conference on Health Informatics
452

Finally, we plan to improve the classification per-
formances by refining some clue parameters in our
proposed method. We specifically refer to the down-
sampling resolution, the Neural Network structure,
and the length of the rhythm pattern.
ACKNOWLEDGMENT
The authors have been supported by the project PON
2014-2020—ARS01 00860 “ATTICUS: Ambient-
intelligent Tele-monitoring and Telemetry for
Incepting and Catering over hUman Sustainability”
funded by the Ministry of Education, University and
Research—RNA/COR 576347.
REFERENCES
Balestrieri, E., Boldi, F., Colavita, A. R., De Vito, L.,
Laudato, G., Oliveto, R., Picariello, F., Rivaldi, S.,
Scalabrino, S., Torchitti, P., et al. (2019). The ar-
chitecture of an innovative smart t-shirt based on the
internet of medical things paradigm. In 2019 IEEE
International Symposium on Medical Measurements
and Applications (MeMeA), pages 1–6. IEEE.
D. Limmer, M. O’Keefe, H. G. B. M. D. B. (2005). Emer-
gency Care Workbook. Pearson.
De Vito, L., Picariello, E., Picariello, F., Tudosa, I., Lo-
previte, L., Avicolli, D., Laudato, G., and Oliveto, R.
(2021). An undershirt for monitoring of multi-lead
ecg and respiration wave signals. In 2021 IEEE Inter-
national Workshop on Metrology for Industry 4.0 &
IoT (MetroInd4. 0&IoT), pages 550–555. IEEE.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
iobank, physiotoolkit, and physionet: components of
a new research resource for complex physiologic sig-
nals. circulation, 101(23):e215–e220.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Islam, S., Ammour, N., and Alajlan, N. (2017). Atrial fib-
rillation detection with multiparametric rr interval fea-
ture and machine learning technique. In 2017 Interna-
tional Conference on Informatics, Health & Technol-
ogy (ICIHT), pages 1–5. IEEE.
Karim, F., Majumdar, S., Darabi, H., and Chen, S. (2017).
Lstm fully convolutional networks for time series clas-
sification. IEEE access, 6:1662–1669.
Kurzweil, R. C., Gibson, L., Albrecht, P., and Grimshaw,
P. (2009). Atrial fibrillation detection. US Patent
7,596,405.
Laudato, G., Boldi, F., Colavita, A. R., Rosa, G., Scal-
abrino, S., Lazich, A., and Oliveto, R. (2020a). Com-
bining rhythmic and morphological ecg features for
automatic detection of atrial fibrillation: Local and
global prediction models. In International Joint Con-
ference on Biomedical Engineering Systems and Tech-
nologies, pages 425–441. Springer.
Laudato, G., Boldi, F., Colavita, A. R., Rosa, G., Scal-
abrino, S., Torchitti, P., Lazich, A., and Oliveto, R.
(2020b). Combining rhythmic and morphological ecg
features for automatic detection of atrial fibrillation.
In HEALTHINF, pages 156–165.
Laudato, G., Scalabrino, S., Colavita, A., Chiacchiari, Q.,
D’Orazio, R., Donadelli, R., De Vito, L., Picariello, F.,
Tudosa, I., Malatesta, R., Gallo, L., and R, O. (2021).
ATTICUS: Ambient-intelligent Tele-monitoring and
Telemetry for Incepting and Catering over hUman
Sustainability. Frontiers in Human Dynamics - Dig-
ital Impacts (Frontiers).
Padmavathi, K. and Ramakrishna, K. S. (2015). Classifi-
cation of ecg signal during atrial fibrillation using au-
toregressive modeling. Procedia Computer Science,
46:53–59.
Pan, J. and Tompkins, W. J. (1985). A real-time qrs de-
tection algorithm. IEEE transactions on biomedical
engineering, (3):230–236.
Sepulveda-Suescun, J., Murillo-Escobar, J., Urda-Benitez,
R., Orrego-Metaute, D., and Orozco-Duque, A.
(2017). Atrial fibrillation detection through heart
rate variability using a machine learning approach
and poincare plot features. In VII Latin American
Congress on Biomedical Engineering CLAIB 2016,
Bucaramanga, Santander, Colombia, October 26th-
28th, 2016, pages 565–568. Springer.
Xiong, Z., Stiles, M. K., and Zhao, J. (2017). Robust ecg
signal classification for detection of atrial fibrillation
using a novel neural network. In 2017 Computing in
Cardiology (CinC), pages 1–4. IEEE.
Xu, S. S., Mak, M.-W., and Cheung, C.-C. (2018). To-
wards end-to-end ecg classification with raw signal
extraction and deep neural networks. IEEE journal of
biomedical and health informatics, 23(4):1574–1584.
Yuan, C., Yan, Y., Zhou, L., Bai, J., and Wang, L. (2016).
Automated atrial fibrillation detection based on deep
learning network. In 2016 IEEE International Con-
ference on Information and Automation (ICIA), pages
1159–1164. IEEE.
Zhou, X., Ding, H., Wu, W., and Zhang, Y. (2015). A real-
time atrial fibrillation detection algorithm based on the
instantaneous state of heart rate. PloS one, 10(9).
Zurro, V., Stelle, A., and Nadal, J. (1995). Detection of
atrial persistent rhythm based on p-wave recognition
and rr interval variability. In Computers in Cardiology
1995, pages 185–188. IEEE.
Simulating the Doctor’s Behaviour: A Preliminary Study on the Identification of Atrial Fibrillation through Combined Analysis of Heart
Rate and Beat Morphology
453
