
Automatic Identification of Non-biting Midges (Chironomidae) using
Object Detection and Deep Learning Techniques
Jack Hollister
1
, Rodrigo Vega
1
and M. A. Hannan Bin Azhar
2
1
School of Psychology and Life Sciences, Canterbury Church University, U.K.
2
School of Engineering, Technology and Design, Canterbury Christ Church University, U.K.
Keywords: Freshwater Ecology, Computer Vison, Object Detection, Image Classification, Chironomidae, Chironomid,
Faster-RCNN, SSD, Raspberry Pi, TensorFlow.
Abstract: This paper introduces an automated method for the identification of chironomid larvae mounted on
microscope slides in the form of a computer-based identification tool using deep learning techniques. Using
images of chironomid head capsules, a series of object detection models were created to classify three genera.
These models were then used to show how pre-training preparation could improve the final performance. The
model comparisons included two object detection frameworks (Faster-RCNN and SSD frameworks), three
balanced image sets (with and without augmentation) and variations of two hyperparameter values (Learning
Rate and Intersection Over Union). All models were reported using mean average precision or mAP. Multiple
runs of each model configuration were carried out to assess statistical significance of the results. The highest
mAP value achieved was 0.751 by Faster-RCNN. Statistical analysis revealed significant differences in mAP
values between the two frameworks. When experimenting with hyperparameter values, the combination of
learning rates and model architectures showed significant relationships. Although all models produced similar
accuracy results (94.4% - 97.8%), the confidence scores varied widely.
1 INTRODUCTION
By measuring the variation in species and their
abundance, biomonitoring assessments can help to
establish the state of an ecosystem (Costa et al.,
2020). It can inform on the quality of water systems,
substrates, or air, and suggest not only what
organisms are present, but what 'should' be present
(Cao et al., 2018). However, these monitoring
systems rely on the correct identification of the
organisms. The two current solutions to this are visual
identification and molecular-based procedures such
as DNA barcoding, but neither is perfect. Visual
methods are prone to mistakes (Haase et al., 2006),
while using DNA barcoding can become incredibly
expensive and time consuming (Shendure et al.,
2017). Using a deep learning based portable platform,
this paper proposes an automated identification
system that is rapid, accurate, cost-effective and
potentially user-friendly.
1
https://orcid.org/0000-0003-4915-9840
2
https://orcid.org/0000-0003-1190-6644
1.1 Freshwater Ecosystems
Freshwater ecosystems can be found on all continents
of the world, but they are most common in North
America, Europe and Asia (Siberia). Only 3% of the
world’s water is fresh water, with majority held
within the polar icecaps (Gerbeaux et al., 2016). A
large portion of living organisms relying on fresh
water as a source of sustenance, and the ecosystems
surrounding these waters provide habitats to a broad
range of species, making it important to maintaining
these systems (Hughes, 2019). There are a range of
fresh waters both natural and manmade such as, but
not limited to, rivers, streams, lakes, marshes, chalk
streams and reservoirs. Despite their importance to
providing sustenance to a large selection of life, and
to supporting the surrounding habitats, freshwater
ecosystems are in danger of degradation due to
anthropogenic interference with the main
contributing factors being pollution, climate change
and habitat transformation (Cao et al., 2018). This
256
Hollister, J., Vega, R. and Azhar, M.
Automatic Identification of Non-biting Midges (Chironomidae) using Object Detection and Deep Learning Techniques.
DOI: 10.5220/0010822800003122
In Proceedings of the 11th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2022), pages 256-263
ISBN: 978-989-758-549-4; ISSN: 2184-4313
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

degradation is having a knock-on effect to the
organisms that depend on these ecosystems. For
instance, the global decrease in macroinvertebrate
populations and the decrease in macroinvertebrate
species diversity is being linked directly to this
anthropogenic interference (Costa et al., 2020),
which, in turn is having a wider knock-on effect to the
ecosystem in which these organisms inhabit (Cao et
al., 2018). To prevent this, ecologists and
conservationists can use biomonitoring techniques to
assess these ecosystems in their current and ongoing
condition. For the biomonitoring of all aquatic
ecosystems, the community structure of benthic
macroinvertebrates can be used and can include the
abundance and presence (or lack of) certain species
(Costa et al. 2020).
Cao et al. (2018) proposed that a lack of expected
benthic macroinvertebrate communities and the
presence of certain ubiquitous species, particularly
those considered pollutant tolerant (i.e., sludge-
worms, Tubifex tubifex), could be used as indicators
to show how the degradation of river water is affected
by municipal waste. This type of approach is
routinely used by researchers and governing bodies
across the globe to assess the quality of water systems
and the surrounding ecosystems. Biggs et al. (2000),
commissioned by the United Kingdom (UK)
Environmental Agency, justified the use of benthic
macroinvertebrates, along with macrophytes and their
presence within different water systems across the
UK, as bioindicators and proposed how the use of
these can be used to assess the water condition of
ponds, lakes and rivers, as well as the condition of the
banks of these water systems. While there is a
selection of species that can contribute to these
assessments (i.e. stone fly nymphs, oligochaetes,
caddisfly larvae), chironomid larvae are considered
ideal candidates for such assessments (Rawal et al.,
2018).
1.2 Chironomids
Chironomids, also known as ‘non-biting midges’ or
‘bloodworms’ (when in their larval stage), are one of
the most abundant and species-rich benthic
macroinvertebrates in freshwater ecosystems
(Nicacio et al., 2015). Chironomids are suggested to
make up 50% of the total benthic macroinvertebrate
population within their respective habitats (Nadjla et
al., 2013). They are found in almost all freshwater
ecosystems including lakes, ponds, swamps, streams
and rivers, and can also be found within isolated
habitats such as tree stumps, and man-made water
ways like flood-prevention drainages. There are an
estimated 600 species found within the United
Kingdom and an estimated 20,000 species worldwide
(Ferrington, 2008). Some species of chironomids can
live in a variety of aquatic systems tolerating a range
of environmental conditions including pH, salinity,
temperature, and sediments, while others require very
specific conditions (Lencioni et al., 2012), and some
can even be found in aquatic systems considered
polluted and inhabitable for most other species
(Luoto, 2011). This has led to the exploration of
chironomids as bioindicators for the general
condition of aquatic ecosystems (Vega et al., 2021),
however, they can also be used for more specific and
streamlined assessments. For instance, Orendt (1999)
created a technical water monitoring method that
provided an acidity assessment for water systems,
where the pH tolerance of 25 species of chironomids
were identified by their presence within several
bodies of water with known pH. Using chironomid
larvae for biomonitoring and paleoclimatic
assessments requires the correct identification to
specific taxonomic levels. However, one of the main
issues with the identification of chironomid larvae is
their minute size. Chironomid larvae are typically
millimetres in length which makes it difficult to
accurately identify them below the taxonomical
classification of family (Chironomidae) without
expert taxonomic knowledge or the means of
molecular-based procedures (Shendure et al., 2017).
1.3 Automated Identification
The use of automatic identification systems is
typically done using computer vison (Azhar et al.,
2012; Ärje et al., 2020), which works with images or
video. These involve techniques such a ‘image
classification’ where a desired subject within an
image can be classified from a set selection of
categories or ‘object detection’, where a desired
subject can be both classified and localised (Rawat et
al., 2017; Huang et al., 2017). The deep learning
techniques, such as Convolutional Neural Network
(CNN), based identifications are growing in
popularity. Bondi et al. (2018) described the
development of an object detection system that uses
techniques from CNNs in order to automatically
detect and identify poachers or high-risk animals in
real-time when used with a video feed. 'PlantSnap' is
another example of integrating deep learning into an
automated identification tool, which can identify and
distinguish over 620,000 different plant species and
their variants from around the world, about 90% of all
described plants (PlantSnap, 2021).
Automatic Identification of Non-biting Midges (Chironomidae) using Object Detection and Deep Learning Techniques
257

There are a number of object detection
frameworks, but two of the more popular ones at
present are Single Shot Detection (SSD) and Faster-
Region-based CNNs (Faster-RCNN) (Arcos-Garcia
et al., 2018; Janahiraman et al., 2019; Bose et al.,
2020). With the object detection system built on top
of CNN, a number of possible models can be used,
including SSD_inception, Faster-RCNN_VGG, and
SSD_ResNet (Zhao et al., 2019). SSD is a framework
for detecting objects first described by Liu et al.
(2016). SSD works in a single step, where the CNN
feeds its learned features to the SSD framework and
then places a grid over an image, with each grid space
including an array of possible default locations,
referred to as anchors or bounding boxes. In SSD,
each grid uses the feature maps from the CNN and
assigns the best anchor to predict objects and their
locations within the image.
Ren et al. (2015) introduced Faster-RCNN as a
two-step approach for object detection, which builds
on a CNN to learn features that are then passed to two
separate functions. One is a regional proposal
network that uses a sliding window approach, but
each window has its own set of anchors. These
anchors will use the feature map to detect any
subjects, but only indicate that there is a subject
within the location and does not define a class for the
location. The second function is the one that defines
the class. These types of deep learning systems
require a training period during which images are fed
into the system, causing the system to learn to
recognise the target within a set of images over time.
Several fine-tuning techniques can be applied to
enhance this training process, including adjusting the
hyperparameters and the quality of the data provided
(Probst et al., 2019; Chudzik et al., 2020).
2 METHODOLOGY
An object detection model designed for three distinct
genera of chironomids (Rheotanytarsus, Cricotopus
and Eukiefferiella) was developed in this
investigation. Two different frameworks were used,
Faster-RCNN (FR) and SSD, in which three different
sets of images (dubbed as A, B and C) were used (255
images, 1500 images and 3000 images respectively).
Following the work of Xia et al. (2018), four learning
rate (LR) hyperparameter values (0.1, 0.001, 0.005,
and 0.0005) were chosen for model performance
comparison. With the optimum LR, three intersection
of union threshold (IOU) values were trailed (0.5, 0.6,
0.7). The IOU threshold is the minimum area allowed
between the overlap of an object detection’s
prediction of where an object is to where it is within
an image with a value between 0 – 1 (Bose et al.,
2020).
Chironomid specimens were collected from the
River Stour in Kent, UK using kick sampling. The
head capsules were mounted on microscope slides
and identified to the genus level. These images were
taken with a Raspberry Pi 3b+ module and a
Raspberry Pi camera v2.1, fitted to a Leica DM 500
high powered microscope. A 4X objective lens was
used for all images (40X total magnification). The
microscope has an internal light source, so no
additional light sources were needed. Microscope
slides containing the mounted specimens of
chironomid larvae were placed on the microscope
stage, secured in place by the stage clips, and images
of the slides were taken. For each microscope slide,
three or four chironomid larvae were mounted with
their head capsules and abdominal segments, and
each specimen was placed under a circular cover slip.
The label on the slide followed the standard labelling
system for specimens mounted on microscope slides
(location, site and date system).
Figure 1: Chironomid larvae head capsules.
Images of two distinct chironomid larvae,
Cricotopus and Eukiefferiella, are shown in Figure 1.
Cricotopus has wide head capsules with thin, curved
mentums, relatively large mandibles and no obvious
antenna. Eukiefferiella has thin head capsules with
dark mandibles and very dark, curved mentums.
Rheotanytarsus has a wide head capsule, mandibles
on the side of their heads, a flat mentum and very
prominent antennae. The mandibles of the three
genera differ in size and shape, so differences in their
shapes and sizes are typically used for morphological
identification and taxonomic classification. The
identified specimens were photographed and the
bounding box labels were applied to the images,
generating a set of co-ordinates for training the object
detection models. An excel reference number was
added so that images could be organized and
referenced easily. Images were separated into their
respective taxonomic group, also known as their
class, at the genus level. There were 863 total images
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
258
taken and used for the three classes (487 Cricotopus,
261 Rheotanytarsus, and 115 Eukiefferiella).
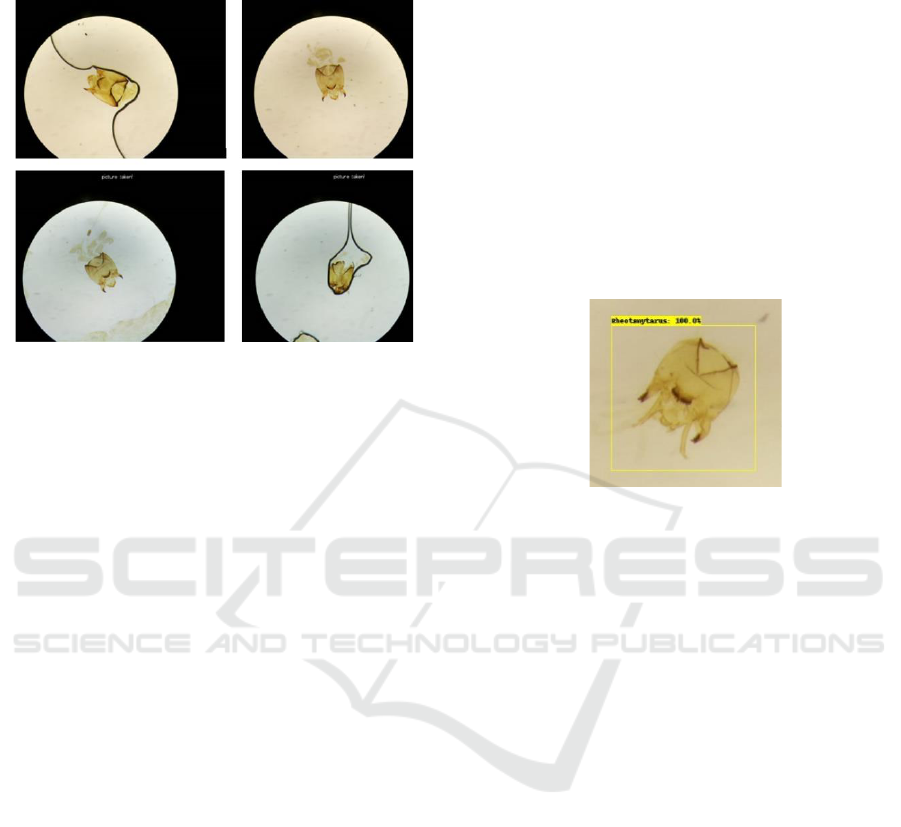
Figure 2: Differences in quality of the images.
Each image contains one chironomid head capsule
of one of three genera, but the quality of the specimen
preparation varies from complete subjects to those
broken apart during mounting procedures or due to
general degradation. Some images contained rear
segments, and some would contain entrails where the
head capsule and rear segments were detached from
each other. Some images also showed wear and
degradation to the slide itself as air and dirt made their
way within the slide. There was also a difference in
the shade of the background on each slide. Figure 2
shows several of the images from the genus
Cricotopus image collection that display differences
in quality, such as the colour of the background, the
completeness of the structure of the head capsule, and
the quality of the slide.
During the training, validation, and testing of the
deep learning models, it was necessary to split the
image sets. To accomplish this, the holdout method
(Yadav et al., 2016) was utilised where a percentage
of the total images was set aside. These images are
taken randomly from the stock of images. In order to
ensure that all models, regardless of image set, could
be evaluated uniformly, 30 images from each class
were set aside for the testing phase. The remaining
images were multiplied to create three image sets (A,
B, and C). In the set A, each class has 85 images.
Thus, to down-sample the majority classes, 85 images
were randomly selected from the original stock within
the Cricotopus and Rheotanytarsus files. In the set B,
each class’s images were up-sampled using
augmentation techniques to create 500 images per
class. For the image set C, images were up-sampled
to 1000 images per class. Several augmentations were
used during the experiments, including rotation to the
left up to 180 degrees, rotation to the right up to 180
degrees, zooming in, zooming out, a horizontal flip,
and a vertical flip. Each image set was split 90:10 for
training and validation respectively.
The training was performed in TensorFlow 1.15,
batch size for each algorithm was 10 and image size
was 300x300. All iterations were run for 5000
epochs. Both object detection frameworks used the
CNN ‘Inception v2’ and the ‘MS COCO’ evaluation
protocols (TensorFlow, 2021). Pretrained models
were downloaded from TensorFlow and used as
transfer learning checkpoints. The mean average
precision or mAP metric was used to evaluate
different object detection models.
Figure 3: Example of a prediction during testing.
Once the models were trained, they were used to
classify specific objects (the chironomid larva head)
from the batch of 90 test images. Figure 3 shows part
of a test image being classified by a model as the
genus Rheotanytarsus, with a confidence score of
100%. A single prediction for each of the test images
was recorded. If there were multiple predictions, only
the prediction with the highest confidence score
would be recorded. Detection thresholds for positive
classification were adjusted to allow all images to be
detected regardless of the confidence value. Using the
confidence scores obtained for all test images, a mean
confidence value was derived for each model.
Additionally, a significance evaluation in the form
of a nested ANOVA (Holmes et al., 2016) was
conducted to examine how different model
configurations were compared, and how image sets
and hyperparameter variations affected the mAP
scores across the trained models. This was then
repeated for the IOU hyperparameter variations while
using the optimum LR for each model configuration
and image set combination. A nested ANOVA can
effectively be broken down into its individual levels
where each could be considered a one-way ANOVA
(Bentler et al., 2010). By following the protocols of a
one-way ANOVA (Doncaster et al., 2007), a
minimum of three repetitions is needed to review the
means of a group for significance, therefore justifying
Automatic Identification of Non-biting Midges (Chironomidae) using Object Detection and Deep Learning Techniques
259
that the number of repetitions of each respective
model chosen was enough to meet the requirements.
3 RESULTS
3.1 Results for Varying LR
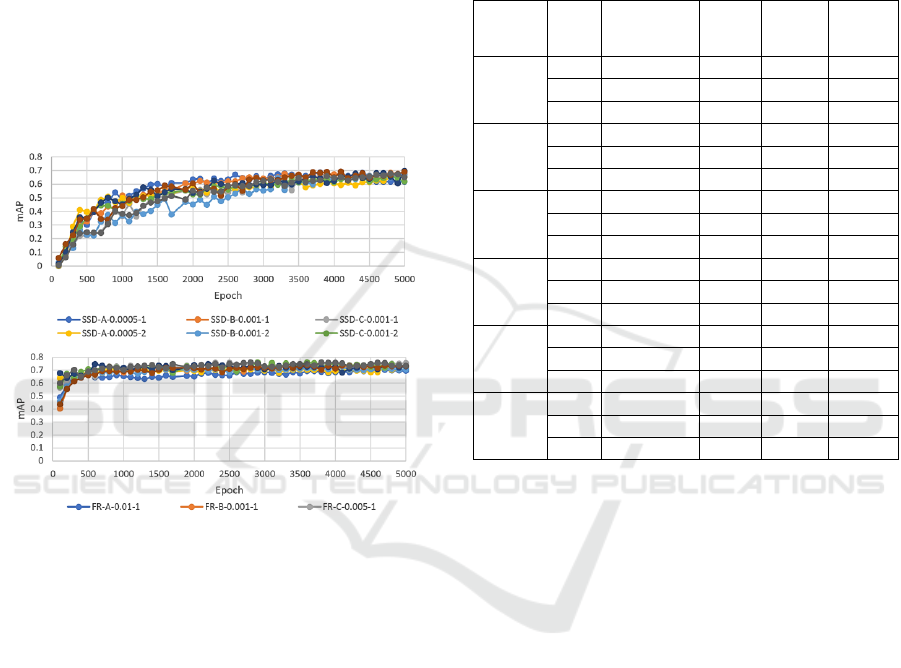
Figure 4 shows how the mAP values developed over
5000 epochs of training with the SSD and Faster-
RCNN model configurations. All SSD models began
with mAP values near zero and gradually increased.
The FR models, however, produced starting mAP
values of over 0.4, increased rapidly, then levelled off
and maintained an mAP value of approximately 0.7.
Figure 4: The mAP values for varying LR.
Among the SSD models, the configuration SSD-
B-0.005 (Model: SSD, image set B and LR 0.005)
produced the highest mAP value of 0.698. Lowest
mAP value obtained for SSD models was 0.507
produced by the SSD-A-0.01 configuration. For the
FR models, the highest mAP was 0.747 obtained by
the configuration FR-A-0.001. The lowest mAP value
obtained for the FR models was 0.624 produced by
the configuration FR-A-0.0005. Averaging the three
runs for each LR, the mean mAP values for SSD
ranged between 0.579 and 0.678, but for FR ranged
between 0.639 and 0.744.
3.2 LR Accuracy and Confidence
Scores
A high accuracy rate was achieved by all models
(95.6%-97.8%). However, confidence scores varied
widely. The configuration FR-B-0.001 at run 2
achieved an average confidence score of 99.9%,
however, all the FR models had an average
confidence score of over 99%. The SSD models, with
the exception of SSD-A-0.0005 at run 2, all achieved
average confidence scores within the range of 80-
90% (Table 1). The lowest average confidence score
of 80.92% was achieved by the model configuration
SSD-C-0.001 at run 3.
Table 1: LR accuracy and confidence scores.
Model
Config.
Run
Accuracy
(%)
Av
Conf
(%)
Min
Conf
(%)
Max
Conf
(%)
SSD-A-
0.0005
1
96.7
83.43
7
100
2
97.8
91.01
3
100
3
97.8
87.64
14
100
SSD-B-
0.001
1
95.6
86.84
19
100
2
97.8
82.95
13
100
3
97.8
89.91
27
100
SSD-C-
0.001
1
97.8
86.10
13
100
2
97.8
86.78
5
100
3
95.6
80.92
15
100
FR-A-
0.01
1
95.6
99.04
71
100
2
96.7
99.56
78
100
3
97.8
99.30
42
100
FR-B-
0.001
1
97.8
99.43
81
100
2
97.8
99.90
93
100
3
97.8
99.80
95
100
FR-C-
0.005
1
97.8
99.74
93
100
2
97.8
99.18
69
100
3
95.6
99.53
84
100
3.3 Significance Evaluations for LR
Based on the analysis of nested ANOVA, it appears
that the mAP values of the two frameworks were
significant and choice of LR affected the model
configurations, but the selection of image sets did not
affect the mAP values. This produced an R
2
value of
81.87%. The nested ANOVA was rerun without the
inclusion of the image sets showing significant
differences in mAP values between the two models
and among the LRs within the models (Model:
F=30.920, SS=0.114, p=0.001, LR: F=3.720,
SS=0.022, p=0.003). This produced an R
2
value of
68.20%. A post-hoc examination (Holmes et al.,
2016) revealed that there was a significant difference
in results between the FR architecture and the
learning rate of 0.0005, and between the SSD
architecture and the learning rate of 0.01 with p-
values <0.05.
3.4 Results for Varying IOUs
Top three model configurations of each framework
(based on the combination of optimum LR values and
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
260
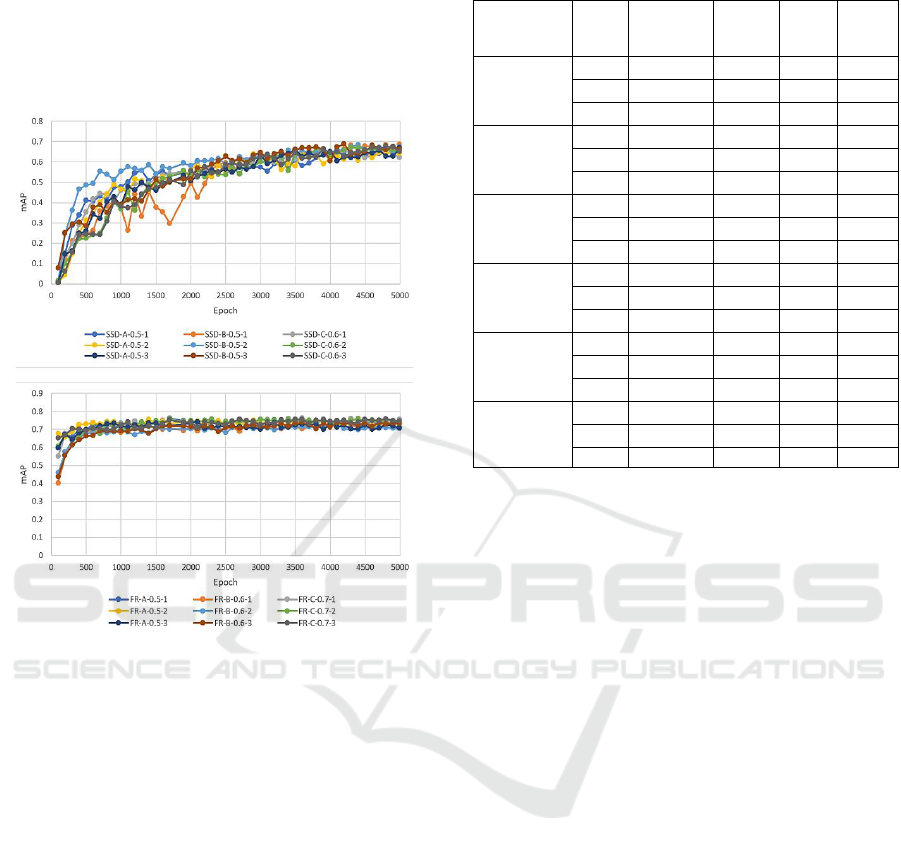
the choice of the image set) were selected to train with
varying IOU settings (0.5, 0.6, 0.7) and each model
was run three times. Averaging the three runs for each
IOU, the mean mAP values for SSD ranged between
0.639 and 0.679, but for the FR models the values
ranged between 0.713 and 0.745.
Figure 5: The mAP values for varying IOUs.
Figure 5 shows how the mAP values developed
during the training over 5000 epochs for the top
performing SSD and FR configurations with varying
IOUs. After 100 epochs, SSD models started with
mAP values near 0 and increased over time, but
remained below 0.7, whereas FR models always
started with values over 0.4, increased rapidly, then
plateaued, and maintained mAP values above 0.7.
3.5 IOU Accuracy and Confidence
Scores
In general, the accuracy scores of all models ranged
from 94.4% to 97.8%, but the confidence scores
differed significantly. The configuration FR-B-0.001-
0.6 (Model: FR, image set B, LR 0.001 and IOU 0.6)
at run 2 achieved the highest average confidence score
of 99.99%, however, all the FR models achieved an
average confidence score above 99% (Table 2). The
configuration SSD-C-0.001-0.6 at run 3 achieved the
lowest average confidence score of 80.92%. The
highest average confidence score for the SSD was
91.98 achieved by SSD-B-0.001-0.5 at run 2.
Table 2: IOU accuracy and confidence scores.
Model
Config
Run
Accuracy
(%)
Av
Conf
(%)
Min
Conf
(%)
Max
Conf
(%)
SSD-A-
0.0005-0.5
1
96.7
89.42
4
100
2
96.7
91.62
13
100
3
96.7
81.00
26
100
SSD-B-
0.001-0.5
1
95.6
86.37
12
100
2
97.8
91.98
39
100
3
96.7
91.17
44
100
SSD-C-
0.001-0.6
1
97.8
86.10
13
100
2
97.8
86.78
5
100
3
95.6
80.92
15
100
FR-A-0.01-
0.5
1
94.4
99.18
64
100
2
96.7
99.46
57
100
3
96.7
99.20
67
100
FR-B- 0.001-
0.6
1
97.8
99.43
81
100
2
97.8
99.99
93
100
3
97.8
99.80
95
100
FR-C-0.005-
0.7
1
97.8
99.62
85
100
2
95.6
99.07
52
100
3
97.8
99.54
69
100
3.6 Significance Evaluations for IOU
The significance of the mAP scores achieved by all
model configurations was evaluated using a nested
ANOVA test by comparing the two frameworks and
how each was affected by the image set and IOUs.
The results show that there were significant
differences between the two frameworks and among
the three image sets: A, B, and C; however, there was
no significant difference among the different IOUs.
This produced an R
2
of 85.09%. After removing IOU
from the nested ANOVA, the results showed
significant differences in mAP values between the
two models and also among different image sets
within the models. (Model: F=29.79, SS=0.067,
p=0.005, Image Set: F=7.18, SS=0.009, p<0.001).
This produced an R
2
of 83.48%. The post-hoc test
revealed that there was a significant difference for FR
with image sets A and C and for SSD with image sets
B and C, all with p-values <0.05.
4 CONCLUSIONS
The intention of this study was to create a cost-
effective and fast-working computer-based model
that could act as an identification tool to aid or replace
more traditional methods such as the visual
identification through morphology or by using
molecular methods of identification. The model can
be executed simply with little computer training, and
Automatic Identification of Non-biting Midges (Chironomidae) using Object Detection and Deep Learning Techniques
261

can do the identification automatically with high
accuracy (>97%). Using the MS COCO metric
system, the model that produced the highest mAP
value (0.751) was the configuration framework FR
using image-set C, LR 0.005 and IOU 0.7. When
comparing the models, the nested ANOVAs showed
significant differences in mAP values between the
SSD and FR frameworks, as expected from previous
studies (Arcos-Garcia et al., 2018; Janahiraman et al.,
2019), however, any significance between the
remaining factors and variables within the model had
not been explored previously. Almost all of the
models using FR achieved mAP values over 0.7 with
the highest reported value of 0.751, whereas the
models using the SSD framework achieved mAP
values under 0.7 with the highest reported value of
0.698. Interestingly, there was very little difference
between any of the models in terms of accuracy. All
models were able to positively classify the majority
of test images with an accuracy of 94.4% - 97.8%.
Previous studies have shown that there is no
universal LR values (Chudzik et al., 2020),
suggesting that each model and its associated neural
network would require an optimisation of its own LR
value. When experimenting with hyperparameter
values, the combination of learning rates and model
architectures showed significant relationships.
Significant effects were found when the SSD
framework was paired with LR 0.01, and when the
FR framework was paired with LR 0.0005. There was
no significant relationship between the different IOU
values trialled and mAP values. However, there was
a small effect of model performance (1.61%
difference in the strength of the relationship with and
without IOU). Thus, all together, varying the IOU
threshold hyperparameter value could be considered
negligible in the general performance output of the
models.
The deep learning method proposed here utilises
trained object detection models and can classify
images in less than a second. In its present state, the
model using object detection and deep learning
involves chironomids to be collected on a site,
euthanised and their head capsules being placed on
microscope slides. These slides are then viewed
through a microscope lens and images are taken.
Images then need to be transferred to a computer
where they can be examined by the object detection
models which will classify the chironomid head
capsule to one of the three genera. The initial stages
require the use of costly workstations and an expert
to work out the optimum training conditions.
However, once the actual model has been developed,
anyone with access to a computer can use it. When
combined with a camera device, such as an affordable
USB camera, this automatic computer model could be
used to identify chironomid larvae specimens just by
passing them in front of a camera feed rather than
using digital images exclusively. However, it is worth
mentioning that this demonstration only covers a very
small fraction of the chironomid diversity, where only
three genera were detected out of an estimated 200+
genera worldwide and did not distinguish species
taxonomy level, where there are an estimated 20,000+
species worldwide. The use of computer vision
models and, in particular, deep learning techniques
for object detection in ecological sciences are still in
their infancy. This study, however, illustrates how
this technique can be used to rapidly identify
taxonomically challenging organisms. It is envisaged
that future work in object detection will open new
opportunities for biological diversity and
biomonitoring, not only of chironomids but also other
group of freshwater organisms.
REFERENCES
Arcos-García, Á., Álvarez-García, J. and Soria-Morillo, L.
(2018). Evaluation of deep neural networks for traffic
sign detection systems. Neurocomputing, 316, 332-344.
Ärje, J, Melvad, C, Jeppesen, MR, et al. (2020) Automatic
image-based identification and biomass estimation of
invertebrates. Methods in Ecol & Evol.; 11: 922– 931.
https://doi.org/10.1111/2041-210X.13428.
Azhar, M.A.H.B., Hoque, S. and Deravi, F. (2012)
Automatic identification of wildlife using Local Binary
Patterns," IET Conference on Image Processing (IPR
2012), pp. 1-6.
Bentler, P. and Satorra, A. (2010). Testing Model Nesting
and Equivalence. Psychological Methods. 15, 111–123.
Biggs, J., Williams, P., Whitfield, M., Fox, G. and Nicolet,
P. (2000). Biological techniques of still water quality
assessment phase 3: method development. Environment
Agency R & D Technical report E110. Environment
Agency. Bristol. UK.
Bondi, E., Fang, F., Hamilton, M., et al. (2018). Spot
poachers in action: Augmenting conservation drones
with automatic detection in near real time. 32nd AAAI
Conf Artif Intell AAAI, 7741-7746.
Bose, S. and Kumar, V. (2020). Efficient inception V2
based deep convolutional neural network for real-time
hand action recognition. IET Img Proc, 14, 688-696.
Cao, X., Chai, L., Jiang, D., et al. (2018). Loss of
biodiversity alters ecosystem function in freshwater
streams: potential evidence from benthic
macroinvertebrates. Ecosphere, 9, e02445.
Chudzik, P., Mitchell, A., Alkaseem, M., et al. (2020).
Mobile real-time grasshopper detection and data
aggregation framework. Sci Rep, 10, 1150.
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
262

Costa, L., Zalmon, I., Fanini, L., et al. (2020).
Macroinvertebrates as indicators of human disturbances
on sandy beaches: A global review. Ecol Indic, 118,
106764.
Doncaster, C., and Davey, A. (2007). Analysis of variance
and covariance: how to choose and construct models
for the life sciences. Cambridge University Press. UK.
Ferrington, L. (2008). Global diversity of non-biting
midges (Chironomidae; Insecta-Diptera) in freshwater.
Hydrobiologia, 595, 447–455.
Gerbeaux, P., Finlayson, C. and van Dam, A. (2016).
Wetland Classification: Overview. In: Finlayson C. et
al. (eds). The Wetland Book. Springer, Dordrecht, The
Netherlands.
Haase, P., Murray-Bligh, J., Lohse, S, et al. (2006).
Assessing the impact of errors in sorting and identifying
macroinvertebrate samples. Hydrobiologia, 566, 505-
521.
Holmes, D., Moody, P., Dine, D., et al. (2016). Research
methods for the biosciences. Oxford University Press,
Oxford, UK.
Huang, J., Rathod, V., Sun, C., et al. (2017).
Speed/accuracy trade-offs for modern convolutional
object detectors. Proc - 30th IEEE Conf Comput Vis Pat
Rec, CVPR, 296-3305.
Hughes, J. (2019). Freshwater ecology and conservation:
Approaches and techniques. Oxford University Press,
Oxford, UK.
Janahiraman, T. and Subuhan, M. (2019). Traffic light
detection using tensorflow object detection framework.
2019 IEEE 9th Int Conf Syst Eng Technol ICSET 2019
- Proceeding. 108-113.
Lencioni, V., Marziali, L. and Rossaro, B. (2012).
Chironomids as bioindicators of environmental quality
in mountain springs. Freshw Sci, 31, 525-541.
Liu, W., Anguelov, D., Erhan, D., et al. (2016). SSD: Single
Shot MultiBox Detector. Comp Vis, 21-37.
Luoto, T. (2011). The relationship between water quality
and chironomid distribution in Finland - A new
assemblage-based tool for assessments of long-term
nutrient dynamics. Ecol Indic. 11, 255-262.
Nadjla, C., Zineb, B., Lilia, F., et al. (2013). Environmental
factors affecting the distribution of chironomid larvae
of the Seybouse wadi, North-Eastern Algeria. J Limnol.
72, 203214.
Nicacio, G. and Juen, L., (2015). Chironomids as indicators
in freshwater ecosystems: An assessment of the
literature. Insect Conserv Divers. 8, 393-403.
Orendt, C. (1999). Chironomids as bioindicators in
acidified streams: A contribution to the acidity
tolerance of chironomid species with a classification in
sensitivity classes. Int Rev Hydrobiol. 84, 439-449.
PlantSnap (2021). PlantSnap website. Available at
https://www.plantsnap.com. Accessed: 1st November
2021.
Probst, P., Boulesteix, A. and Bischl, B. (2019). Tunability:
Importance of hyperparameters of machine learning
algorithms. J Mach Learn Res. 20, 1-22.
Rawal, D., Verma, H. and Prajapat, G. (2018). Ecological
and economic importance of Chironomids (Diptera).
JETIR, vol. 5.
Rawat W and Wang Z. (2017). Deep Convolutional Neural
Networks for Image Classification: A Comprehensive
Review. Neural Comput. 29, 2352-2449.
Ren, S., He, K., Girshick, R. and Sun, J. (2015). Faster R-
CNN: towards real-time object detection with region
proposal networks. In Proceedings of the 28th
International Conference on Neural Information
Processing Systems - Volume 1 (NIPS'15).
Shendure, J., Balasubramanian, S., Church, G., et al.
(2017). DNA sequencing at 40: Past, present and future.
Nature. 550, 345-353.
Tensorflow (2021), TensorFlow GitHub Model Zoo.
Available at: https://github.com/tensorflow/models/
blob/master/research/object_detection/g3doc/tf1_detec
tion_zoo.md , Accessed: 1st November 2021.
Vega, R, Brooks, S.J., Hockaday, W., et al. (2021).
Diversity of Chironomidae (Diptera) breeding in the
Great Stour, Kent: baseline results from the Westgate
Parks Non-biting Midge Project. J Nat Hist. 55, 11-12.
Xia, D., Chen, P., Wang, B., et al.(2018). Insect Detection
and Classification Based on an Improved
Convolutional Neural Network. Sensors
(Basel).18(12):4169. doi:10.3390/s18124169
Yadav, S., and Shukla, S. (2016). Analysis of k-Fold Cross-
Validation over Hold-Out Validation on Colossal
Datasets for Quality Classification. IACC, 78-83.
Zhao, Z.Q., Zheng, P., Xu, S.T. and Wu, X., (2019). Object
detection with deep learning: A review. IEEE Trans
Neural Netw Learn Syst, 30, 3212-3232.
Automatic Identification of Non-biting Midges (Chironomidae) using Object Detection and Deep Learning Techniques
263
