
Efficient Removal of Weak Associations in Consensus Clustering
N. C. Ruckiya Sinorina, Howard J. Hamilton
a
and Sandra Zilles
b
Department of Computer Science, University of Regina, 3737 Wascana Parkway, Regina, SK, Canada
Keywords:
Consensus Clustering, Association Matrix.
Abstract:
Consensus clustering methods measure the strength of an association between two data objects based on how
often the objects are grouped together by the base clusterings. However, incorporating weak associations in the
consensus process can have a negative effect on the quality of the aggregated clustering. This paper presents
an efficient automatic approach for removing weak associations during the consensus process. We compare
our approach to a brute force method used in an existing consensus function, NegMM, which tends to be rather
inefficient in terms of runtime. Our empirical analysis on multiple datasets shows that the proposed approach
produces consensus clusterings that are comparable in quality to the ones produced by the original NegMM
method, yet at a much lower computational cost.
1 INTRODUCTION
Consensus clustering refers to the process of generat-
ing an aggregation or ensemble of several individual
clusterings of a dataset, called base clusterings. Typ-
ically, in consensus clustering the final clusters are
based on how frequently the data objects are grouped
together in the base clusterings (Strehl and Ghosh,
2003). Such techniques have been used in a broad
range of applications, including image processing,
network analysis, business process management, and
cloud computing (Wu et al., 2018). Other than cre-
ating robust and high-quality final clusters, consensus
clustering supports distributed data mining, data pri-
vacy (Chalamalla, 2010), and knowledge re-usability.
To find a common ground of agreement, some
consensus approaches analyze the association of data
objects to each other (Fred and Jain, 2005; Strehl and
Ghosh, 2003; Zhong et al., 2019). An association in-
dicates that a pair of data objects occur together in
a cluster, in some or all input base clusterings. Some
consensus functions examine each data object pair for
associations and form a pairwise association matrix
to generate the final clusterings (Fred and Jain, 2005;
Strehl and Ghosh, 2003; Zhong et al., 2019). The val-
ues in an association matrix range from 0 to 1 based
on the frequency with which a data object pair is clus-
tered together in the base clusterings. An association
is weak if the frequency of a pair of data objects to oc-
a
https://orcid.org/0000-0003-1475-0980
b
https://orcid.org/0000-0001-7834-8574
cur in the same cluster is low, and the majority of base
clusterings keep the two objects in separate clusters.
Several consensus functions use association matrices
without distinguishing between strong and weak asso-
ciations; instead they treat each non-zero association
value as an association to take into account in cal-
culating a final consensus clustering (Fred and Jain,
2005; Strehl and Ghosh, 2003).
Recently it was shown that removing weak asso-
ciations during consensus clustering can improve the
quality of the result (Baller et al., 2018; Zhong et al.,
2019). But how can one make a clear distinction
of weak and strong pairwise associations in the base
clusterings? In other words, which threshold value in
the interval from 0 to 1 should be the largest associ-
ation value below which the associations are consid-
ered weak? We call such value a weak association
threshold, and the problem of choosing it the thresh-
old selection problem. It is usually hard to answer
the above question in advance, and the best threshold
value will in general depend on the dataset and the
base clusterings (Baller et al., 2018).
To the best of our knowledge, the only existing
consensus method that attempts to solve the threshold
selection problem is NegMM (Zhong et al., 2019). It
is a brute force method that tests about 50 threshold
values and generates consensus candidates for all of
them. One of these consensus candidates is then cho-
sen as the final output. Since NegMM searches for the
best weak association threshold among a large num-
ber of candidates, each time invoking the consensus
326
Sinorina, N., Hamilton, H. and Zilles, S.
Efficient Removal of Weak Associations in Consensus Clustering.
DOI: 10.5220/0010820800003116
In Proceedings of the 14th International Conference on Agents and Artificial Intelligence (ICAART 2022) - Volume 3, pages 326-335
ISBN: 978-989-758-547-0; ISSN: 2184-433X
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

mechanism, it is computationally expensive.
In this paper, we address the question whether it
is possible to select a weak association threshold au-
tomatically based on the distribution of values in the
association matrix for a set of base clusterings con-
sidered for consensus, without exhaustively probing a
large number of thresholds. We propose an approach,
called the WAT (Weak Association Threshold) ap-
proach, to solve the threshold selection problem by
computing a threshold value based on the distribu-
tion of association values. In particular, our approach
is to apply a standard clustering algorithm (e.g., K-
Means (Aggarwal and Reddy, 2014) or Gaussian Mix-
ture Model clustering (Reynolds, 2015)) on the set of
association values, group the latter into two clusters,
and use the threshold value that separates these two
clusters as our weak association threshold.
In an empirical analysis on 16 datasets, we com-
pare the original NegMM method to two variants that
use our WAT approach in place of the brute force
threshold search. Our observations strongly suggest
that similar quality clusterings can be obtained with
much reduced computational cost when using the
WAT approach instead of generating numerous con-
sensus candidates in the original NegMM consensus
function.
2 RELATED WORK
Several procedures for aggregating clusterings for a
dataset have been proposed over the years. This pa-
per primarily focuses on consensus functions based
on pairwise associations. To combine the base clus-
terings and find the aggregated result, such consensus
functions measure the similarity of two data points or
clusters in the base clusterings (Vega-Pons and Ruiz-
Shulcloper, 2011). To measure this similarity, these
methods consider the incidence of the pair grouped
in the same cluster by different clustering methods.
A convenient and effective representation of the pair-
wise data similarity measure is the association matrix
(Fred and Jain, 2005). With data points as rows and
columns, the values in this association matrix indi-
cate how many times a pair of objects are grouped in
the same cluster in the given base clusterings relative
to the total number of base clusterings considered for
consensus.
Fred and Jain proposed generating consensus re-
sults based on an association matrix using the Evi-
dence Accumulation method (EAC). It applies single-
linkage agglomerative clustering over an association
matrix of base clusterings to generate the consensus
(Fred and Jain, 2005).
Such an association matrix is a summary repre-
sentation of the participating base clusterings. As the
consensus function runs, evidence is accumulated in
the association matrix and the quality of the final con-
sensus depends on the information in this representa-
tion. For example, the existence of many uncertain
data pairs in an association matrix can affect the fi-
nal consensus. Uncertain data pairs are pairs that are
clustered together in about half of the base clusterings
and separated in the other half.
The problem of uncertain data pairs is addressed
in (Yi et al., 2012), by proposing the creation of a par-
tially observed association matrix, with reliable val-
ues only for the data pairs whose cluster memberships
are agreed upon by most of the clustering algorithms.
A matrix completion algorithm completes the matrix
and then applies spectral clustering to generate the fi-
nal consensus. This approach defines a lower bound
of 0.2 and an upper bound of 0.8 for an association
value to be treated as uncertain. These thresholds may
not be ideal across all datasets and base clusterings.
Recently the NegMM consensus was proposed,
which removes weak associations from the associa-
tion matrix (Zhong et al., 2019). Evidence that occurs
in lower frequency in base clusterings is considered
as negative information, and treated as noise in the
association matrix. Including this negative evidence
between data objects is demonstrated to cause unde-
sirable effects in the consensus and degrade the con-
sensus performance (Zhong et al., 2019). Removing
weak associations means to decrease the frequency of
such undesired pairs in the association matrix. The
authors recommend removing small similarity values,
where the threshold for a weak association value lies
in the range from 0.01 to 0.5. NegMM consensus con-
siders all the values in this range, with an increment
of 0.01, as potential weak association thresholds. The
basic idea of NegMM consensus is to generate sev-
eral candidate consensus clusterings by removing the
weak associations using each possible threshold value
one at a time. The best clustering among the candi-
dates is then selected using a clustering internal va-
lidity measure. The details are discussed in Section 3.
The NegMM consensus function relies on a pre-
defined set of thresholds to remove weak associations
during consensus. In our work, the main distinctive
feature is to deploy methods that can select a weak
association threshold without evaluating an extensive
set of candidates. We propose two ways of select-
ing weak association thresholds and explore which
method performs well under which conditions.
Efficient Removal of Weak Associations in Consensus Clustering
327

Input : Base clusterings for dataset with n
objects, number of clusters K
Output: Final clustering C
∗
Generate an n ×n pairwise association matrix
of base clusterings, AM;
Initialize Candidates =
/
0,
thresholds = {0,0.01,0.02,...,0.5};
for τ ∈thresholds do
AM
∗
= AM;
if AM
∗
i j
≤ τ then
AM
∗
i j
= 0;
end
C
τ
= Ncut(AM
∗
,K);
Candidates = Candidates ∪C
τ
;
end
Evaluate MM index for each
C
τ
∈Candidates;
Return C
∗
= C
τ
where C
τ
∈Candidates
minimizes MM index;
Algorithm 1: Algorithm for NegMM consensus.
3 NegMM CONSENSUS
NegMM consensus is the “Negative Evidence Re-
moved Clustering Ensemble”, in which associations
of data pairs with low co-occurrence frequency in the
base clusterings are removed from the association ma-
trix (Zhong et al., 2019). Algorithm 1 displays the
method for NegMM consensus. It initially generates
a data-based association matrix using the input base
clusterings (line 1). For a dataset with n objects, using
z base clusterings P = {P
1
,..,P
z
}, an n×n data-based
association matrix AM is generated using
AM
i j
= α(x
i
,x
j
)/z (1)
where α(x
i
,x
j
) is the number of times x
i
and x
j
oc-
cur together in the same cluster, across all base clus-
terings in P. NegMM considers all data pairs with
association values at most 0.5 as potentially weak as-
sociations. Weakly associated data points may appear
in two different clusters of the original base cluster-
ings, and such associations can be negative evidence
for consensus. However, as it is not easy to decide
which association evidence is negative information in
the matrix and ought to be removed, NegMM gradu-
ally removes association evidence within the range of
0 to 0.5, with an increase of 0.01 in each step. All as-
sociation values below the selected value in each step
are set as zeros to generate a modified association ma-
trix (lines 4 to 7). Then, normalized cut (Ncut) (Shi
and Malik, 2000) is applied over each of these mu-
tated association matrices, to generate candidate con-
sensus clusterings (line 8). Ncut is a graph partition-
ing algorithm – often applied for instance in image
segmentation – that uses a normalized cut criterion to
form the partitions, where the associations of edges
within the partitions of the graph are used for normal-
izing the cut. The reader is referred to (Shi and Malik,
2000) for more details on Ncut.
Among these candidate consensus clusterings, the
highest quality one is selected as the final output clus-
tering, using a minimax similarity-based index (MM
index, lines 11 to 12). The MM index is an inter-
nal validity index to assess the quality of a clustering
(Zhong et al., 2019), that can be calculated without
accessing the original dataset. It defines a good clus-
tering as one in which the clusters have high stability
and low cohesion with other clusters. For a clustering
C = {C
1
,C
2
,...,C
K
} with K clusters, the MM index is
defined as follows.
MM =
K
∑
i=1
coh(C
i
,X \C
i
)/stb(C
i
) (2)
where coh(C
i
,C
j
) is the cohesion between clusters C
i
and C
j
and stb(C
i
) is the stability of cluster C
i
in terms
of density-based connectivity (Zhong et al., 2019).
Cohesion and stability are given by
coh(C
i
,X \C
i
) = max
x
a
∈C
i
,x
b
∈X\C
i
RS(x
a
,x
b
,S
X
,l)(3)
stb(C
i
) = min
x
a
∈C
i1
,x
b
∈C
i2
RS(x
a
,x
b
,S
C
i
,l) (4)
where “\” denotes the set difference, and C
i1
and C
i2
are created by bi-partitioning C
i
, here using Ncut. For
an undirected graph G(X,E) over a data set X, with
similarity matrix S
X
, RS is the robust minimax sim-
ilarity value. This robust path-based minimax simi-
larity measure for x
a
and x
b
with l as the number of
nearest neighbours is defined as
RS(x
a
,x
b
,S
X
,l) (5)
= max
p∈ρ
X
ab
{ min
1≤h<|p|
sim(p[h], p[h + 1])w
h
w
h+1
} (6)
where ρ
X
ab
is set of all paths between x
a
and x
b
, p[h] is
the h
th
vertex in path p, sim(x
a
,x
b
) is the similarity of
x
a
and x
b
from S
X
, and w
h
is the weight for x
h
(Zhong
et al., 2019).
Increased stability indicates a high within-cluster
density and low cohesion indicates a low density-
based connectivity to other clusters. The MM index
favours clusterings that correspond to high-density re-
gions separated by low-density regions (Zhong et al.,
2019).
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
328

Input : Base clusterings for dataset with n
objects
Output: n ×n pairwise association matrix
SM
∗
without weak associations
Generate n ×n pairwise association matrix of
base clusterings, SM. Initialize i = 0, j = 0;
Let X be a sorted array of values in SM
greater than zero;
Generate two clusters in X using a clustering
algorithm;
select the maximum value in the left cluster
as τ;
while i < n and j < n do
if SM
i j
≤ τ then
SM
∗
i j
= 0;
else
SM
∗
i j
= SM
i j
;
end
end
Algorithm 2: Algorithm to remove weak associations.
4 APPROACH
Figure 1 gives an overview of the consensus process.
Our proposed WAT approach towards the threshold
selection problem is to apply a standard clustering al-
gorithm on the association values, thus cluster the as-
sociation values into two clusters, and use this two-
way clustering to derive a weak association threshold
τ. This paper tests the WAT approach with two clus-
tering techniques that are applied to the association
values, namely K-Means and Gaussian Mixture Mod-
els (GMM), to select the threshold τ automatically.
Algorithm 2 gives the general method. Initially,
one generates the pairwise association matrix of com-
ponent pairs in the base clusterings. With a dataset of
n objects, Algorithm 2 generates an n ×n data-based
association matrix SM (line 1). It then groups the as-
sociation values in the matrix into two clusters using
a basic clustering algorithm. These two clusters sep-
arate the larger association values in the matrix from
the smaller ones. The cluster that contains the mini-
mum association value that is greater than zero is re-
ferred to as the left cluster, and the cluster that con-
tains the maximum association value is referred to as
the right cluster. The highest value in the left cluster
is the cut-off value for the association value groups
and is the weak association threshold τ (line 4).
In our experiments, we tested two methods for
obtaining the required grouping of association values
into two clusters :
• K-Means with K = 2
• Gaussian Mixture Model Clustering using the EM
algorithm to model the set of association values as
a mixture of two Gaussians.
One sets all the values less than or equal to the
selected threshold to zero in the pairwise association
matrix (lines 5 to 10). The consensus is then gen-
erated using the new association matrix without the
weak associations. See Figure 2 for the K-Means
clusters and the Gaussians for the histogram of associ-
ation values for a synthetic dataset called Jain (Fr
¨
anti
and Sieranoja, 2018).
The proposed way of removing the weak associa-
tions can be used with any consensus function that op-
erates on an association matrix, such as, for example
NegMM or CSPA (Strehl and Ghosh, 2003). This pa-
per analyzes the approach empirically in the context
of the NegMM consensus function. Instead of gen-
erating several consensus candidates, using the WAT
approach in the NegMM consensus function means to
first calculate the threshold to remove weak associa-
tions and then to perform the consensus only with the
derived weak association threshold.
5 EMPIRICAL ANALYSIS
5.1 Experimental Datasets and
Implementation Details
The basic details of 16 datasets used in our exper-
iments, both real-world and synthetic, are given in
Table 1. The Blobs dataset with isotropic Gaussian
blobs for clustering analysis is generated using the
make blobs function with default parameters in Scikit
learn (Pedregosa et al., 2011). For evaluation pur-
poses, the datasets are all classification datasets with
ground truth labels. The experiments are run on a
Windows OS computer with an Intel(R) Xeon(R) E3
processor, running at 3.70 GHz, and 8 GB of RAM.
All implementations are done in Python 3.7.
We experimented with four different sets of base
clusterings per dataset. In three of these four setups,
the mechanism for generating base clusterings is us-
ing multiple initializations of the K-Means clustering
algorithm. The experiments in this paper use the K-
Means implementation in Scikit-learn with parame-
ters n clusters, the number of clusters K set as de-
scribed below, max iter, the maximum number itera-
tions set to 4 instead of the default value of 300, and
n init, the number of restarts set to 1 instead of the de-
fault value of 10 (Pedregosa et al., 2011). Small val-
ues are used for max iter and the number of restarts to
increase the diversity of the clusterings produced by
Efficient Removal of Weak Associations in Consensus Clustering
329
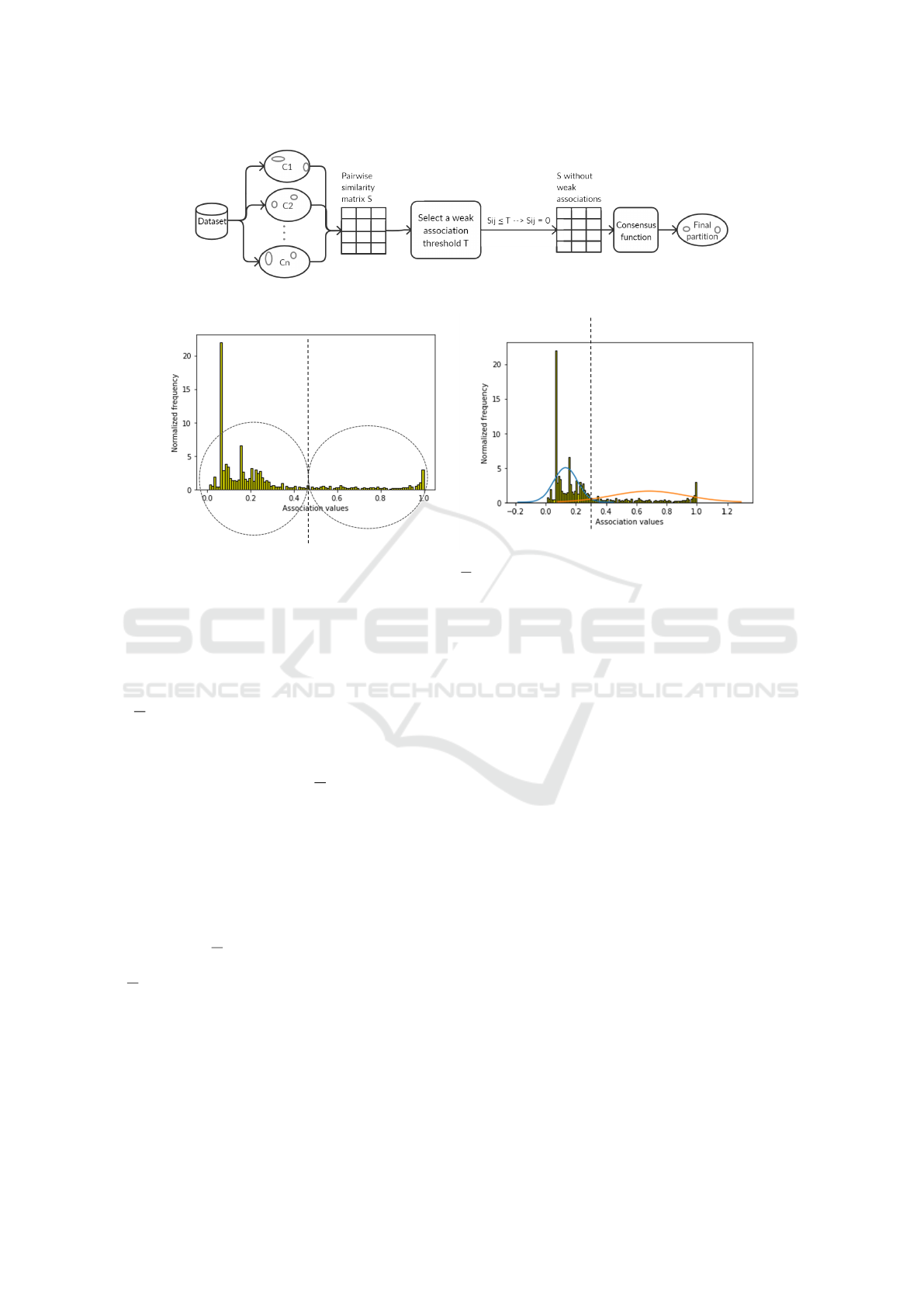
Figure 1: General overview of consensus clustering with removal of weak associations.
Figure 2: Histograms of data point association values obtained using 100 base clusterings on the Jain dataset. The base
clusterings were formed by K-Means clustering with K =
√
N and number of iterations set to 4. The dotted vertical line
separates the association values into two clusters, as determined by 2-Means (left) and GMM (right). The weak association
thresholds are 0.46 (left) and 0.33 (right).
the multiple initializations. All other parameters used
are defaults.
Setup 1: 1000 Base Clusterings, K-Means with K =
b
√
Nc. To create similar experimental settings as in
the original article on NegMM (Zhong et al., 2019),
a set of 1000 base clusterings is generated for each
dataset where K-Means is run 1000 times, with the
number of clusters K fixed as b
√
Nc, for a dataset with
N instances.
Setup 2 (Resp. 3): 100 (Resp. 10) Base Clusterings,
K-Means with Random K. To investigate the effect
of using the proposed WAT approach over different
consensus sizes, this paper tests consensus functions
using 100 and 10 base clusterings separately, for all
datasets. Here, K-Means is run with random K rang-
ing from 2 to
√
N with the assumption that the appro-
priate number of clusters in a dataset will be at most
√
N (Bezdek and Pal, 1998).
Setup 4: 10 Diverse Base Clusterings. More di-
verse base clusterings are expected to be obtained by
using a variety of clustering algorithms to generate the
base clusterings. We generate 10 base clusterings for
each dataset by using 10 methods: (1) K-Means (Ag-
garwal and Reddy, 2014), (2) Mini Batch K-Means
(Sculley, 2010), (3) the density based clustering algo-
rithm DBSCAN (Aggarwal and Reddy, 2014), (4-6)
hierarchical agglomerative clustering (Aggarwal and
Reddy, 2014) with its three linkage variations Ward,
Complete, and Average (Pedregosa et al., 2011), (7)
Mean Shift (Comaniciu and Meer, 2002), (8) BIRCH
(Zhang et al., 1997), (9) Gaussian Mixture (Aggarwal
and Reddy, 2014), and (10) Bayesian Gaussian Mix-
ture (Roberts et al., 1998).
The evaluation measure used to compare the clus-
terings are Adjusted Rand Index (ARI) (Hubert and
Arabie, 1985) and Adjusted Mutual Index (AMI)
(Vinh et al., 2010). The purpose of the WAT approach
is to remove weak associations in a substantially more
efficient way than NegMM does, without suffering a
substantial loss of quality in the resulting consensus
clusterings. Therefore, we also recorded runtimes.
Below we use NegMM-WAT(K) (NegMM-
WAT(GMM), resp.) to denote the version of NegMM
when replacing the brute force threshold search by
our WAT method using K-Means with K=2 (using
GMM with 2 Gaussians resp.)
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
330

Table 1: Basic details of the datasets used in our analysis where attributes define the dimensionality of the dataset.
Dataset #Objects #Attributes #Classes Type
Flame (Fr
¨
anti and Sieranoja, 2018) 240 2 2 Synthetic
Path Based (PB) (Fr
¨
anti and Sieranoja, 2018) 300 2 3 Synthetic
Jain (Fr
¨
anti and Sieranoja, 2018) 373 2 2 Synthetic
Compound (CM) (Fr
¨
anti and Sieranoja, 2018) 399 2 6 Synthetic
R15 (Fr
¨
anti and Sieranoja, 2018) 600 2 15 Synthetic
Blobs 650 2 4 Synthetic
AG (Fr
¨
anti and Sieranoja, 2018) 788 2 7 Synthetic
Iris (Dua and Graff, 2017) 150 4 3 Real
Wine (Dua and Graff, 2017) 178 13 3 Real
Image Segmentation (IS) (Dua and Graff, 2017) 210 19 7 Real
Seeds (Dua and Graff, 2017) 210 7 3 Real
Glass (Dua and Graff, 2017) 214 9 6 Real
User Knowledge Model (UKM) (Dua and Graff, 2017) 258 5 4 Real
Ecoli (Dua and Graff, 2017) 336 8 8 Real
Libras Movement (LM) (Dua and Graff, 2017) 360 90 15 Real
Yeast (Dua and Graff, 2017) 1484 8 10 Real
5.2 Experimental Results
Tables 2–5 compare the consensus functions
NegMM, NegMM-WAT(K), and NegMM-
WAT(GMM) in terms of ARI (left) and AMI
(right). For the WAT methods, the percentage of
increase or decrease compared to NegMM is shown
in parentheses. Avg Base refers to the average ARI or
AMI of the base clusterings, with standard deviation
in parentheses.
With 1000 K-Means base clusterings, using the
same setup as in the literature, all three consensus
functions, NegMM, NegMM-WAT(K), and NegMM-
WAT(GMM), produced clusterings of similar quality
in most cases. The final clustering performance is ei-
ther the same or slightly improved in terms of ARI
and AMI values for NegMM-WAT(K) method, ex-
cept for the AG and IS datasets. For the AG dataset,
in this case, the AMI results are identical too. Now,
considering the results for the NegMM-WAT(GMM),
again the performance is the same or slightly im-
proved compared to that of the original NegMM, for
all datasets other than AG, where WAT(GMM) causes
a very small loss in quality.
This raises the question of whether similar obser-
vations can be made for more diverse sets of base
clusterings than those that were tested in (Zhong et al.,
2019). Our additional experiments using 100 and 10
base clusterings adopt the same experimental setup
except for setting the number K of clusters for K-
Means base clusterings randomly between 2 and
√
N.
Varying the number of clusters in base clusterings in-
creases the diversity of the base clusterings.
Consensus results of 100 base clusterings in
this case, using NegMM, NegMM-WAT(K), and
NegMM-WAT(GMM), are given in Table 3. For the
Jain, R15, Iris, Wine, and Glass datasets, the ARI and
AMI values for all three consensus functions are the
same. For the Flame, PB, and Blobs datasets, using
the WAT methods on top of NegM gave slightly im-
proved values, but the differences were minor.
For the Seeds dataset, NegMM with WAT(K) has
the same result as NegMM, but, using NegMM with
WAT(GMM) improves the clustering by 190% and
68% for ARI and AMI, respectively. This is be-
cause the threshold selected by WAT(GMM) is 0.05,
whereas NegMM and WAT(K) selected the thresholds
0.4 and 0.44, respectively (see Figure 3). Though
NegMM’s best candidate clustering with the small-
est MM index is for threshold 0.4, the ARI and AMI
evaluations suggest that this clustering is inferior to
the clustering for threshold 0.05, in this case.
For the IS and CM datasets, the original NegMM
method outperformed the WAT variants in terms of
ARI and AMI. Possibly the distribution of association
values relates better to a clustering into more than two
groups, rather than one with two groups (as assumed
by our WAT approach). For example, see the asso-
ciation values of the 100 base clusterings in the IS
dataset, given in Figure 3. For the AG dataset, while
using WAT(GMM) decreases the clustering perfor-
mance, NegMM and NegMM-WAT(K) produce con-
sensus clusterings of the same quality.
When generating 10 base clusterings using K-
Means with independently randomly chosen values
for K, the consensus functions NegMM and NegMM-
WAT(K) and NegMM-WAT(GMM) produced compa-
rable results for the datasets Flame, CM, Iris, Wine,
Ecoli, and Yeast. The clustering quality improved
by using WAT methods for the UKM dataset and
Efficient Removal of Weak Associations in Consensus Clustering
331

Table 2: Consensus results over 1000 base clusterings obtained from K-Means with fixed K. The consensus functions
NegMM, NegMM-WAT(K), and NegMM-WAT(GMM) are compared in terms of ARI (left) and AMI (right).
ARI AMI
Dataset NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base
Flame 0.45 0.45 (0%) 0.45 (0%) 0.45 (0.02) 0.4 0.4 (0%) 0.40 (0%) 0.40 (0.01)
PB 0.46 0.46 (0%) 0.46 (0%) 0.43 (0.05) 0.55 0.55 (0%) 0.55 (0%) 0.51 (0.05)
Jain 0.32 0.32 (0%) 0.32 (0%) 0.32 (0.01) 0.37 0.37 (0%) 0.37 (0%) 0.37 (0.01)
CM 0.54 0.54 (0%) 0.54 (0%) 0.54 (0.07) 0.72 0.72 (0%) 0.72 (0%) 0.71 (0.04)
R15 1 1.00 (0%) 1.00 (0%) 1.00 (0.00) 1 1.00 (0%) 1.00 (0%) 1.00 (0.00)
Blobs 0.82 0.82 (0%) 0.82 (0%) 0.82 (0.00) 0.84 0.84 (0%) 0.84 (0%) 0.84 (0.00)
AG 0.79 0.76 (-4%) 0.75 (-5%) 0.72 (0.04) 0.88 0.88 (0%) 0.87 (-1%) 0.84 (0.03)
Iris 0.72 0.73 (1%) 0.73 (1%) 0.73 (0.05) 0.74 0.76 (3%) 0.76 (3%) 0.75 (0.03)
Wine 0.37 0.37 (0%) 0.37 (0%) 0.36 (0.01) 0.43 0.43 (0%) 0.43 (0%) 0.43 (0.01)
IS 0.51 0.45 (-12%) 0.51 (0%) 0.40 (0.03) 0.66 0.65 (-2%) 0.66 (0%) 0.57 (0.03)
Seeds 0.72 0.72 (0%) 0.72 (0%) 0.71 (0.04) 0.69 0.69 (0%) 0.69 (0%) 0.70 (0.03)
Glass 0.54 0.55 (2%) 0.54 (0%) 0.49 (0.05) 0.74 0.74 (0%) 0.74 (0%) 0.70 (0.04)
UKM 0.17 0.17 (0%) 0.17 (0%) 0.19 (0.05) 0.26 0.27 (4%) 0.26 (0%) 0.26 (0.07)
Ecoli 0.42 0.42 (0%) 0.42 (0%) 0.43 (0.06) 0.62 0.62 (0%) 0.62 (0%) 0.60 (0.03)
LM 0.34 0.34 (0%) 0.34 (0%) 0.31 (0.02) 0.62 0.62 (0%) 0.62 (0%) 0.59 (0.01)
Yeast 0.13 0.13 (0%) 0.13 (0%) 0.13 (0.02) 0.26 0.25 (-4%) 0.26 (0%) 0.25 (0.02)
Figure 3: Plots for the distribution of NegMM-based association values for the Seeds, IS, and CM datasets with 100 base
clusterings. Different separators in the plot are where NegMM, NegMM-WAT(K), and NegMM-WAT(GMM), resp., put their
thresholds.
Figure 4: Distribution of NegMM-based association values of the Seeds, Jain, and R15 datasets with 10 K-Means base
clusterings. Different separators in the plot indicate where NegMM, NegMM-WAT(K), and NegMM-WAT(GMM) put their
thresholds, resp.
yielded slightly improved performance for the PB
and LM datasets, in terms of ARI and AMI values.
The WAT(K) method in this case for the Blobs and
Glass datasets also is comparable in performance to
NegMM. Using the NegMM-WAT(GMM) method,
for the dataset Seeds one obtains an improvement in
ARI by 21% and in AMI by 10%. The various thresh-
olds selected by the three consensus functions for the
Seeds dataset in this case, as shown in Figure 4, sug-
gests that WAT(GMM) selected the better threshold.
For both the Jain and the R15 dataset, from several
candidate clusterings, NegMM selected the clustering
with threshold 0 as its final output and obtained per-
fect clusters (ARI and AMI are 1). That is, the con-
sensus of 10 random base clusterings for Jain and R15
identified the exact clusters in the datasets without re-
moving any associations. However, the WAT methods
selected thresholds near 0.5, as shown in Figure 4, for
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
332
Table 3: Consensus results over 100 base clusterings obtained from K-Means with random K. The consensus functions
NegMM, NegMM-WAT(K), and NegMM-WAT(GMM) are compared in terms of ARI (left) and AMI (right).
ARI AMI
Dataset NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base
Flame 0.93 0.95 (2%) 0.97 (4%) 0.23 (0.13) 0.88 0.90 (2%) 0.93 (6%) 0.49 (0.04)
PB 0.49 0.50 (2%) 0.50 (2%) 0.34 (0.09) 0.57 0.57 (0%) 0.57 (0%) 0.55 (0.04)
Jain 1 1.00 (0%) 1.00 (0%) 0.15 (0.07) 1 1.00 (0%) 1.00 (0%) 0.46 (0.04)
CM 0.64 0.48 (-25%) 0.54 (-16%) 0.40 (0.11) 0.8 0.70 (-13%) 0.72 (-10%) 0.70 (0.04)
R15 1 1.00 (0%) 1.00 (0%) 0.66 (0.27) 1 1.00 (0%) 1.00 (0%) 0.87 (0.12)
Blobs 0.8 0.82 (2%) 0.82 (2%) 0.37 (0.19) 0.83 0.84 (1%) 0.84 (1%) 0.65 (0.08)
AG 0.98 0.98 (0%) 0.78 (-20%) 0.43 (0.18) 0.98 0.98 (0%) 0.90 (-8%) 0.76 (0.07)
Iris 0.57 0.57 (0%) 0.57 (0%) 0.46 (0.13) 0.71 0.71 (0%) 0.71 (0%) 0.67 (0.04)
Wine 0.37 0.37 (0%) 0.37 (0%) 0.36 (0.01) 0.43 0.43 (0%) 0.43 (0%) 0.43 (0.01)
IS 0.46 0.40 (-13%) 0.40 (-13%) 0.32 (0.13) 0.59 0.58 (-2%) 0.58 (-2%) 0.52 (0.14)
Seeds 0.2 0.20 (0%) 0.58 (190%) 0.38 (0.13) 0.38 0.38 (0%) 0.64 (68%) 0.57 (0.04)
Glass 0.55 0.55 (0%) 0.55 (0%) 0.41 (0.14) 0.74 0.74 (0%) 0.74 (0%) 0.67 (0.08)
UKM 0.17 0.17 (0%) 0.17 (0%) 0.19 (0.06) 0.26 0.27 (4%) 0.26 (0%) 0.26 (0.07)
Ecoli 0.42 0.42 (0%) 0.4 (-5%) 0.42 (0.05) 0.62 0.62 (0%) 0.6 (-3%) 0.60 (0.02)
LM 0.32 0.32 (0%) 0.32 (0%) 0.23 (0.09) 0.6 0.60 (0%) 0.61 (2%) 0.48 (0.14)
Yeast 0.13 0.13 (0%) 0.13 (0%) 0.12 (0.02) 0.26 0.25 (-4%) 0.26 (0%) 0.25 (0.02)
Table 4: Consensus results over 10 base clusterings obtained from K-Means with random K. The consensus functions
NegMM, NegMM-WAT(K), and NegMM-WAT(GMM) are compared in terms of ARI (left) and AMI (right).
ARI AMI
Dataset NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base
Flame 0.97 0.95 (-2%) 0.95 (-2%) 0.30 (0.14) 0.93 0.90 (-3%) 0.90 (-3%) 0.47 (0.06)
PB 0.44 0.47 (7%) 0.52 (18%) 0.37 (0.09) 0.53 0.52 (-2%) 0.59 (11%) 0.57 (0.04)
Jain 1 0.08 (-92%) 0.72 (-28%) 0.13 (0.05) 1 0.25 (-75%) 0.64 (-36%) 0.49 (0.04)
CM 0.54 0.54 (0%) 0.53 (-2%) 0.40 (0.08) 0.76 0.76 (0%) 0.76 (0%) 0.70 (0.03)
R15 1 0.79 (-21%) 0.83 (-17%) 0.63 (0.33) 1 0.93 (-7%) 0.90 (-10%) 0.84 (0.17)
Blobs 0.82 0.82 (0%) 0.64 (-22%) 0.43 (0.21) 0.84 0.84 (0%) 0.71 (-15%) 0.68 (0.09)
AG 0.99 0.92 (-7%) 0.72 (-27%) 0.38 (0.13) 0.99 0.93 (-6%) 0.87 (-12%) 0.77 (0.05)
Iris 0.57 0.57 (0%) 0.57 (0%) 0.50 (0.13) 0.71 0.71 (0%) 0.71 (0%) 0.68 (0.04)
Wine 0.37 0.37 (0%) 0.37 (0%) 0.37 (0.01) 0.43 0.43 (0%) 0.43 (0%) 0.43 (0.01)
IS 0.44 0.38 (-14%) 0.39 (-11%) 0.33 (0.13) 0.6 0.57 (-5%) 0.57 (-5%) 0.55 (0.08)
Seeds
0.48 0.43 (-10%) 0.58 (21%) 0.40 (0.16) 0.58 0.45 (-22%) 0.64 (10%) 0.58 (0.05)
Glass 0.53 0.54 (2%) 0.49 (-8%) 0.35 (0.1) 0.72 0.73 (1%) 0.69 (-4%) 0.66 (0.09)
UKM 0.18 0.26 (44%) 0.26 (44%) 0.23 (0.06) 0.26 0.32 (23%) 0.32 (23%) 0.30 (0.07)
Ecoli 0.42 0.43 (2%) 0.43 (2%) 0.44 (0.06) 0.6 0.6 (0%) 0.6 (0%) 0.59 (0.03)
LM 0.29 0.3.00 (3%) 0.31 (7%) 0.20 (0.11) 0.58 0.58 (0%) 0.59 (2%) 0.44 (0.17)
Yeast 0.14 0.13 (-7%) 0.14 (0%) 0.12 (0.03) 0.26 0.26 (0%) 0.26 (0%) 0.25 (0.03)
the same set of base clusterings, and the clustering
quality is not as good as that obtained by NegMM.
The last analysis uses 10 diverse base clusterings
obtained from multiple clustering algorithms. A con-
sensus over these base clusterings is again formed
using NegMM and NegMM-WAT(K). ARI and AMI
values of the final clusterings are given in Table 5.
For most of the datasets, the NegMM-WAT(K)
consensus of these diverse 10 base clusterings gives
final clusterings whose ARI and AMI values are com-
parable to those obtained by the original NegMM
method. For the Iris dataset, the AMI is unchanged,
but the ARI declines by 19%. Similarly, compar-
ing the WAT(GMM) results over those of the original
NegMM, for many datasets, the differences seemed
to be negligible. Interestingly, for the Glass dataset,
the clustering quality in terms of ARI improved by
37%, and for CM datasets by 9%. Also, the cluster-
ing quality of the Yeast data set improved using both
WAT methods in terms of ARI and AMI. In case of
Wine data set the threshold NegMM was able to pick
better threshold than WAT methods.
The runtime comparison of NegMM and its WAT
variants are shown in Figure 5. The NegMM consen-
sus involves a one-time generation of an association
matrix, multiple consensus steps to create candidate
clusterings, and selecting the best candidate as the fi-
nal clustering. By contrast, the WAT approach run-
Efficient Removal of Weak Associations in Consensus Clustering
333
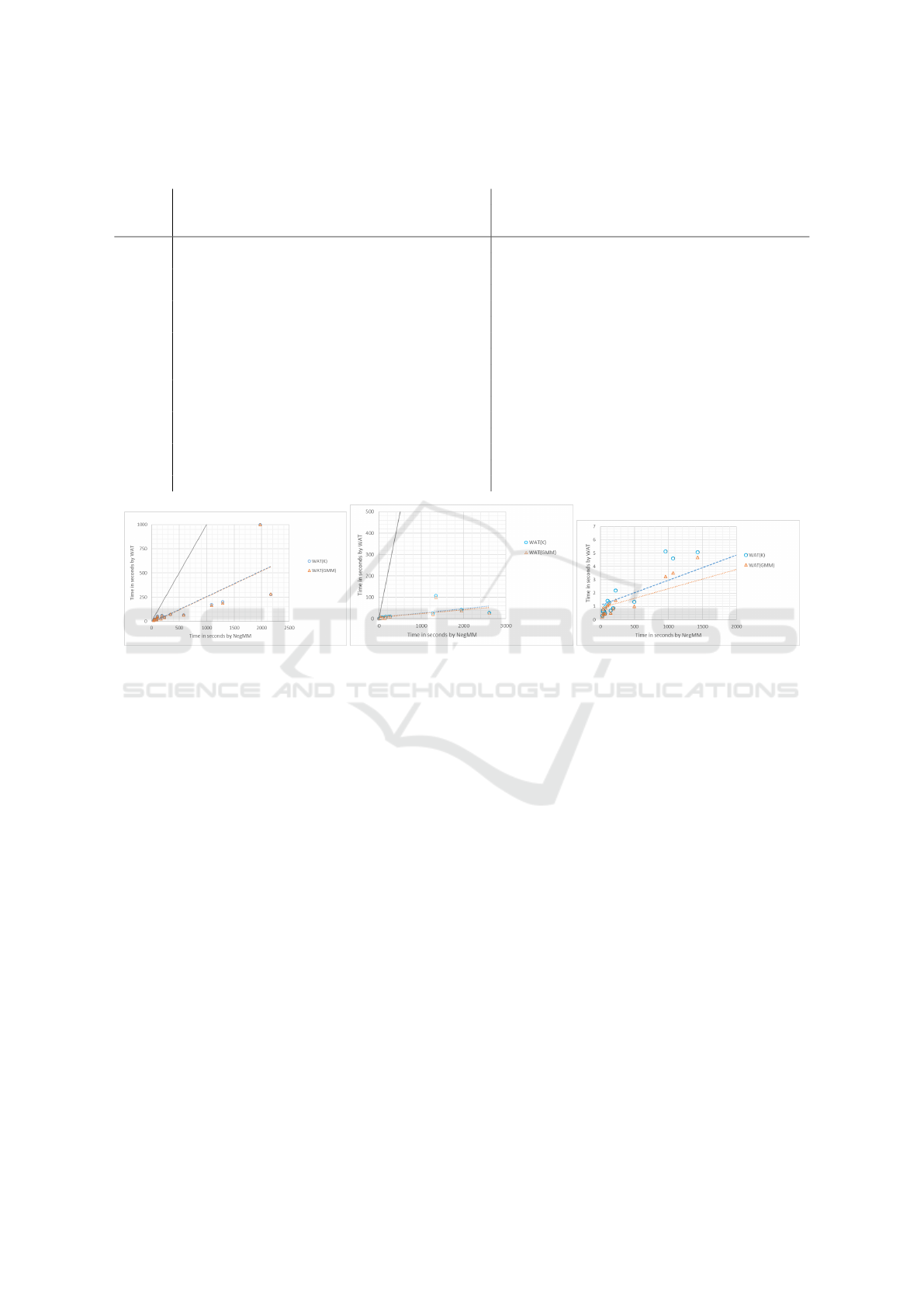
Table 5: Consensus results over 10 diverse base clusterings obtained from 10 clustering algorithms. The consensus functions
NegMM, NegMM-WAT(K), and NegMM-WAT(GMM) are compared in terms of ARI (left) and AMI (right).
ARI AMI
Dataset NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base NegMM
NegMM
-WAT(K)
NegMM
-WAT(GMM)
Avg Base
Flame 0.45 0.45 (0%) 0.45 (0%) 0.27 (0.19) 0.46 0.46 (0%) 0.46 (0%) 0.32 (0.19)
PB 0.48 0.48 (0%) 0.48 (0%) 0.32 (0.22) 0.56 0.56 (0%) 0.56 (0%) 0.40 (0.23)
Jain 0.56 0.56 (0%) 0.56 (0%) 0.39 (0.36) 0.54 0.54 (0%) 0.54 (0%) 0.45 (0.27)
CM 0.57 0.57 (0%) 0.62 (9%) 0.63 (0.11) 0.71 0.71 (0%) 0.77 (8%) 0.74 (0.06)
R15 1 1.00 (0%) 1.00 (0%) 0.78 (0.27) 1 1.00 (0%) 1.00 (0%) 0.94 (0.08)
Blobs 0.81 0.81 (0%) 0.81 (0%) 0.75 (0.05) 0.83 0.83 (0%) 0.84 (1%) 0.85 (0.02)
AG 0.78 0.83 (6%) 0.80 (3%) 0.81 (0.07) 0.88 0.91 (3%) 0.88 (0%) 0.88 (0.04)
Iris 0.69 0.56 (-19%) 0.59 (-14%) 0.59 (0.20) 0.74 0.74 (0%) 0.62 (-16%) 0.69 (0.10)
Wine 0.91 0.55 (-40%) 0.55 (-40%) 0.55 (0.40) 0.89 0.68 (-24%) 0.68 (-24%) 0.62 (0.33)
IS 0.48 0.47 (-2%) 0.46 (-4%) 0.28 (0.20) 0.62 0.63 (2%) 0.61 (-2%) 0.50 (0.21)
Seeds 0.81 0.81 (0%) 0.81 (0%) 0.54 (0.35) 0.76 0.76 (0%) 0.76 (0%) 0.58 (0.27)
Glass 0.19 0.19 (0%) 0.26 (37%) 0.17 (0.14) 0.38 0.35 (-8%) 0.35 (-8%) 0.36 (0.14)
UKM 0.27 0.27 (0%) 0.27 (0%) 0.14 (0.13) 0.34 0.33 (-3%) 0.33 (-3%) 0.25 (0.15)
Ecoli 0.48 0.49 (2%) 0.47 (-2%) 0.38 (0.26) 0.61 0.63 (3%) 0.6 (-2%) 0.51 (0.16)
LM 0.31 0.31 (0%) 0.31 (0%) 0.19 (0.14) 0.6 0.59 (-2%) 0.59 (-2%) 0.48 (0.21)
Yeast 0.16 0.17 (6%) 0.17 (6%) 0.06 (0.07) 0.26 0.29 (12%) 0.28 (8%) 0.22 (0.12)
Figure 5: Runtime comparison of NegMM to NegMM-WAT(K), and NegMM to NegMM-WAT(GMM), using 1000, 100, and
10 base clusterings for datasets. Each point refers to one dataset. The dotted lines give the best fits for the recorded data. The
solid line is the graph of y = x. In the rightmost graph the line for y = x is not shown, since with the chosen scales it would be
nearly identical to the y-axis, indicating a substantial runtime advantage of WAT over NegMM.
time involves creating the association matrix, select-
ing the threshold from the distribution, and the gener-
ation of a single consensus clustering. The trend line
in the scatter plot is the best fit for the run-time data,
and it generalizes the observations.
The slope of the trend line for 1000 base cluster-
ings is 0.129, which means that NegMM consensus
takes about 7.7 times as much time as its WAT vari-
ants using 1000 base clusterings. Generally, the time
to create an association matrix is quadratic in the size
of the dataset and linear in the number of base clus-
terings used. With 1000 base clusterings, a consid-
erable portion of runtime for both the NegMM and
WAT variants is utilized for generating the matrix it-
self. Despite this, the rate of runtime difference is
noticeable.
The runtime plot for 100 base clusterings has trend
lines of even smaller slopes (around 0.012) compared
to that for 1000 base clusterings. The average ratio
indicates that NegMM takes about 51.4 times as much
time as its WAT variants using 100 base clusterings.
Using 10 base clusterings, on average NegMM takes
233 times as much time as its WAT variants.
Based on these results, we conclude that our WAT
approach yields noticeable runtime savings without
substantially reducing the clustering quality when
compared to NegMM in the experimental setting that
was originally tested in (Zhong et al., 2019). The run-
time savings increase with smaller consensus sizes,
while the clustering quality stays comparable to that
of NegMM, with a tendency to higher losses for
smaller consensus sizes.
6 CONCLUSION
The NegMM consensus function relies on removing
weak associations from the association matrix to im-
prove the final clustering. This consensus function
is effective and outperforms other consensus function
to create high quality clusterings for many datasets
(Zhong et al., 2019). However, to select the best clus-
tering, NegMM generates several candidate cluster-
ICAART 2022 - 14th International Conference on Agents and Artificial Intelligence
334

ings, partially removing associations each time us-
ing a range of threshold values. The WAT approach
in combination with NegMM instead determines one
threshold, and generates the final consensus directly.
Our empirical results for the majority of datasets over
different generation mechanisms and varied consen-
sus sizes suggest that the WAT approach is success-
ful in removing the weak associations to attain simi-
lar quality clusterings in much-reduced runtime com-
pared to the original NegMM method. This was evi-
dent in particular in our experiments with 1000 and
with 100 base clusterings. Further studies will be
needed to determine why the WAT approach hurt
the clustering performance of NegMM more in the
case of 10 less diverse base clusterings. Moreover,
the WAT approach surprisingly improved the qual-
ity of some NegMM clusterings, for example, apply-
ing WAT(GMM) for the consensus of 100 and 10 K-
Means base clusterings for the Seeds dataset.
ACKNOWLEDGEMENTS
Funding for this research was provided by ISM
Canada and the Natural Sciences and Engineering Re-
search Council of Canada.
REFERENCES
Aggarwal, C. C. and Reddy, C. K. (2014). Data Cluster-
ing: Algorithms and Applications. CRC Press, Boca
Raton, Florida, USA.
Baller, T., Hamilton, H., and Zilles, S. (2018). A meta ap-
proach to removing weak links during consensus clus-
tering. Unpublished manuscript.
Bezdek, J. C. and Pal, N. R. (1998). Some new indexes of
cluster validity. IEEE Transactions on Systems, Man
& Cybernetics. Part B (Cybernetics), 28(3):301–315.
Chalamalla, A. K. (2010). A survey on consensus cluster-
ing techniques. Technical report, Department of Com-
puter Science, University of Waterloo.
Comaniciu, D. and Meer, P. (2002). Mean shift: A robust
approach toward feature space analysis. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
24(5):603–619.
Dua, D. and Graff, C. (2017). UCI machine learning repos-
itory. University of California, Irvine, School of Infor-
mation and Computer Sciences.
Fr
¨
anti, P. and Sieranoja, S. (2018). K-means properties
on six clustering benchmark datasets. Applied Intel-
ligence, 48(12):4743–4759.
Fred, A. L. and Jain, A. K. (2005). Combining multiple
clusterings using evidence accumulation. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
27(6):835–850.
Hubert, L. and Arabie, P. (1985). Comparing partitions.
Journal of Classification, 2(1):193–218.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., Vanderplas, J., Passos,
A., Cournapeau, D., Brucher, M., Perrot, M., and
Duchesnay, E. (2011). Scikit-learn: Machine learning
in Python. Journal of Machine Learning Research,
12(85):2825–2830.
Reynolds, D. A. (2015). Gaussian mixture models. In Li,
S. Z. and Jain, A. K., editors, Encyclopedia of Biomet-
rics, pages 827–832. Springer, Boston, MA, USA.
Roberts, S. J., Husmeier, D., Rezek, I., and Penny, W.
(1998). Bayesian approaches to Gaussian mixture
modeling. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 20(11):1133–1142.
Sculley, D. (2010). Web-scale K-means clustering. In
Proceedings of the 19th International Conference on
World Wide Web (WWW 2010), pages 1177–1178.
Shi, J. and Malik, J. (2000). Normalized cuts and image
segmentation. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 22(8):888–905.
Strehl, A. and Ghosh, J. (2003). Cluster ensembles: A
knowledge reuse framework for combining multiple
partitions. Journal of Machine Learning Research,
3:583–617.
Vega-Pons, S. and Ruiz-Shulcloper, J. (2011). A sur-
vey of clustering ensemble algorithms. Intl. Jour-
nal of Pattern Recognition and Artificial Intelligence,
25(3):337–372.
Vinh, N., Epps, J., and Bailey, J. (2010). Informa-
tion theoretic measures for clusterings comparison:
Variants, properties, normalization and correction for
chance. Journal of Machine Learning Research,
11(2010):2837–2854.
Wu, X., Ma, T., Cao, J., Tian, Y., and Alabdulkarim, A.
(2018). A comparative study of clustering ensem-
ble algorithms. Computers & Electrical Engineering,
68:603–615.
Yi, J., Yang, T., Jin, R., Jain, A. K., and Mahdavi, M.
(2012). Robust ensemble clustering by matrix com-
pletion. In Proceedings of the 12th IEEE International
Conference on Data Mining, (ICDM 2012), pages
1176–1181.
Zhang, T., Ramakrishnan, R., and Livny, M. (1997).
BIRCH: A new data clustering algorithm and its ap-
plications. Data Min. Knowl. Discov., 1(2):141–182.
Zhong, C., Hu, L., Yue, X., Luo, T., Qiang, F., and Haiyong,
X. (2019). Ensemble clustering based on evidence ex-
tracted from the co-association matrix. Pattern Recog-
nition, 92:93–106.
Efficient Removal of Weak Associations in Consensus Clustering
335
