
Coordinate Attention UNet
Quoc An Dang and Duc Dung Nguyen
Computer Science and Engineering Faculty, Ho Chi Minh City University of Technology, Vietnam
Keywords:
Brain Tumor Segmentation, Instant Segmentation, Channel Attention, Coordinate Attention.
Abstract:
In this paper, we propose an alternative architecture based on the UNet, which utilized the attention module.
Our model solved the context loss and feature dilution caused by sampling operation of the UNet model using
the enhancement ability of the attention. Further more, we applied one of the latest attention module named
Coordinate Attention module to our model and proposed modification of this module to improve the effective
of this module for Magnetic Resonance Imaging (MRI) scans.
1 INTRODUCTION
Gliomas brain tumor is the most aggressive malig-
nant primary brain tumor. It mostly occur in adults
having low survival rate (Tamimi and Juweid, 2017).
For diagnosing the tumor, The traditional method is
segmenting the Magnetic Resonance Imaging (MRI)
by specialist, which is very costly and time consum-
ing. Therefore the needed of an automated segment
method arisen.
In recent years, many researchers working on the
method to segmenting brain tumor. The work varied
from the basic CNN model (Havaei et al., 2015) to the
encoder-decoder architecture like UNet (Ronneberger
et al., 2015), UNet++ (Zhou et al., 2018), VNet (Mil-
letari et al., 2016), nnUNet (Isensee et al., 2018).
Then in order to utilize the z-axis feature, many 3D
model appeared like 3D UNet (C¸ ic¸ek et al., 2016), 3D
Dilated Multi-Fiber Network(Chen et al., 2019), 3D
autoencoder regularization (Myronenko, 2018). And
among the lots of research, UNet model still appeared
to be one of the most typical baseline model. How-
ever this model still have problem with the context
loss and feature dilution. In this paper, we propose
a new model to address this problem of UNet model
by utilizing one of the latest attention module named
Coordinate attention (Hou et al., 2021).
Along side the development of baseline model,
attention modules have also been proved to achieve
high results in brain tumor segmentation including
Multi-scale guided attention (Sinha and Dolz, 2019),
Cross-task Guided Attention (Zhou et al., 2019) and
Attention UNet (Oktay et al., 2018). Furthermore,
many attention modules have been proved very ef-
fective in segmentation task such as Squeeze-and-
Excitation (Hu et al., 2017) and Coordinate atten-
tion (Hou et al., 2021). Even though these attentions
have high results in segmenting normal image. Their
method is not suitable to work with MRI scans which
have high variance. We also propose a modification
of coordinate attention to cope with the MRI scans.
In order to demonstrate the effective of our de-
signed. We did experiments on the BraTS 2020
dataset (Menze et al., 2015) with the origin UNet
model and our proposal. The results show the im-
provement of our proposal in both model design and
attention module design.
2 RELATED WORK
In this section, we present a brief overview of recent
method to handle the context problem of UNet model
and attention module design.
2.1 UNet Model Improvement
Many work have tried to solve the context problem of
UNet model. Attention U-Net (Oktay et al., 2018)
applied attention gate to enhance the skip connec-
tion features. UNet++ (Zhou et al., 2018) redesigned
the skip connections to reduce the gap between con-
tracting path and expanding path. UNet 3+ (Huang
et al., 2020) applied the full-scale skip connection to
incorporate low-level details with high-level seman-
tics from feature maps in different scales. DC-UNet
(Lou et al., 2021) proposed the dual channel block to
replace the traditional convolution blocks.
122
Dang, Q. and Nguyen, D.
Coordinate Attention UNet.
DOI: 10.5220/0010657700003061
In Proceedings of the 2nd International Conference on Robotics, Computer Vision and Intelligent Systems (ROBOVIS 2021), pages 122-127
ISBN: 978-989-758-537-1
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
2.2 Attention Modules
Attention have been proved effective in brain tumor
segment task. Using attention gate to filter the skip
connection in Attention UNet (Oktay et al., 2018),
applying attention at multi-scale feature in Multi-
scale guided attention (Sinha and Dolz, 2019), Cross-
task Guided Attention (Zhou et al., 2019) devel-
oped a specific module to work with multi tasking
model. Squeeze-and-Excitation Network (Hu et al.,
2017) proposed a attention module to captured the
channel-wise relationship. Inspired by the Squeeze-
and-Excitation Network, coordinate attention (Hou
et al., 2021) further encoding coordinate info along
with channel-wise relationship.
3 METHOD
In this section, we propose the design of UNet model
with attention module in the first part. In the second
part, we propose a modification of the Coordinate At-
tention module (Hou et al., 2021) to be more suitable
with MRI scans data.
3.1 Coordinate Attention UNet
In 2015, UNet model (Ronneberger et al., 2015) is
proposed, which achieved the highest ranking in ISBI
cell tracking challenge in the same year. The model is
divided into two parts. A contracting path to capture
context and a symmetric expanding path that enables
precise localization.
Both paths are comprised of multiple convolution
layers followed by sampling operations. The con-
tracting path executed downsampling, which reduc-
ing the size of image. On the contrary, the expanding
path used upsampling to expanded the encoded fea-
tures causing the feature dilution. To further enhance
the context for upsampling operations, UNet model
added skip connections between contracting path and
expanding path to use the context from the contract-
ing to enhance the diluted features.
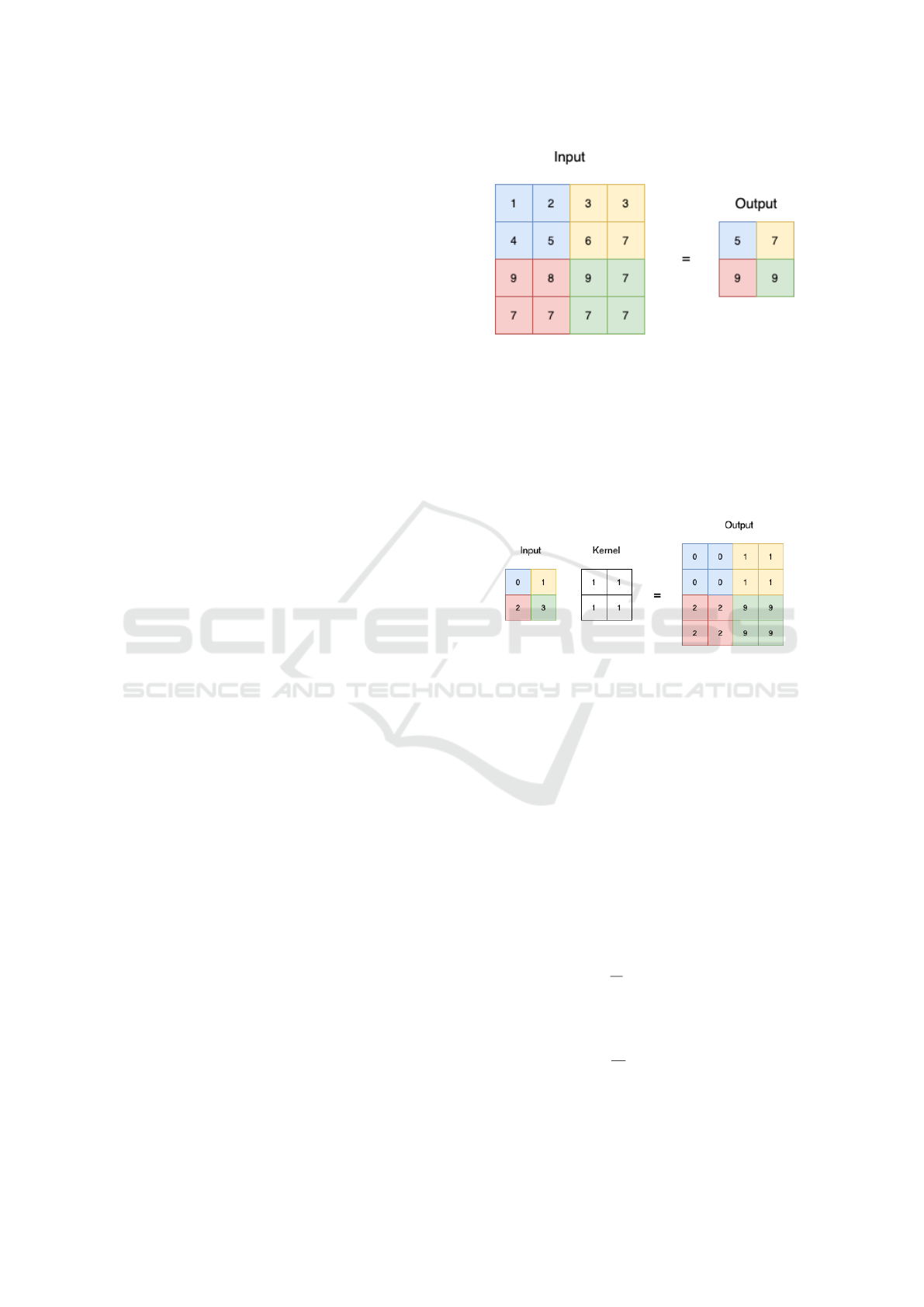
Downsampling step is done by applying 2×2 max
pooling. Figure 1 show a sample result of this oper-
ation. As we can see, the size of the input is reduced
by half, which meaned half of the feature is lost after
downsampling. In other to utilize the lost features, we
applied coordinate attention to the feature map before
passing it to the downsampling. This result in the de-
sign of the left side of our model showed in Figure
3.
Upsampling is done by applying transposed con-
volution with kernel size 2 × 2 and stride 2. Fig-
Figure 1: Max pooling oparation sample with kernel 2 × 2
and stride 2.
ure 2 demonstrate a sample result of this operation.
As we can see, the encoded features are expanded
by two times. By applying coordinate attention to
the expanded segment map, we augmented it with
direction-aware and position-sensitive attention maps.
We demonstrated this design in the right side of Fig-
ure 3.
Figure 2: Transposed convolution of input size 2 × 2 and
kernel size 2 × 2.
3.2 Coordinate Attention for MRI Scan
Coordinate attention module (Hou et al., 2021) can
be divided into two part. The first part is called co-
ordinate information embedding and the second part
is coordinate information generation. In this work we
modified the the first part of the module.
Developed from Squeeze-and-Excitation attention
(Hu et al., 2017), Coordinate Attention (Hou et al.,
2021) have added the ability to encode coordinate at-
tention by applying average pooling for along hori-
zontal coordinate and vertical coordinate. Given the
input X, the output of the c-th channel at width w is
formulated as:
z
w
c
=
1
H
∑
0≤ j<H
x
c
( j, w) (1)
And the output of the c-th channel at height h is for-
mulated as:
z
h
c
=
1
W
∑
0≤i<W
x
c
(h, i) (2)
While this method have been proven effective, it
is not suitable to use average pooling for data high
Coordinate Attention UNet
123

Figure 3: Coordinate Attention UNet architecture.
variance (Boureau et al., 2010) such as MRI scans.
Therefore we changed the pooling function to max
pooling. The c-th channel output at width w become:
z
w
c
= max
0≤ j<H
x
c
( j, w) (3)
And the output of the c-th channel at height h is
formulated as:
z
h
c
= max
0≤i<W
x
c
(h, i) (4)
Then the encoded features is then passed through
attention generation part to generate two attention fea-
tures. These features is used to re-weight the input
features.
4 EXPERIMENT
In this section, we describe the dataset we use to eval-
uate our models. Next we explain how we setup the
experiments and evaluate the output. Finally we show
the comparison result of our model.
4.1 BraTS Dataset
In our experiment, we use BraTS 2020 dataset
(Menze et al., 2015) for training our model. This
dataset is used for holding 2020 BraTS Challenge
(Bakas et al., 2018), which is the most famous chal-
lenge in Brain Tumor segmentation task. The data
provided is collected from real patients and labeling
by specialist.
The data is provided as a 3D image in niffty for-
mat. One image contains 155 layers of 240 × 240 im-
ages. Further more, the intensity value of images is
not in the range of 0-255 like normal images, which
make the data more special to handle.
Data of a single patients includes four modalities
of MRI scans, native image (T1), a post-contrast T1-
weighted (T1Gd), T2-weighted (T2), and T2 Fluid
Attenuated Inversion Recovery (T2-FLAIR). Figure
4 show a sample of each image types.
The dataset have three segment labels, the GD-
enhancing tumor (ET — label 4), the peritumoral
edema (ED — label 2), and the necrotic and non-
enhancing tumor core (NCR/NET — label 1).
4.2 Experiment Setup
In order to evaluate our work, we implement three
models: original UNet model, Coordinate Attention
UNet model (CA-UNet), Max pooling Coordinate At-
tention UNet model (MCA-UNet).
All model is implemented using PyTorch (Paszke
et al., 2019). We use Adam Optimizer with the learn-
ing rate 1 × 10
−5
. Model is trained using the batch
size of 5 in 5 epochs. As the attention module is
harder to converge, we first train the UNet model then
used the pretrained weight to training CA-UNet and
MCA-UNet. The result is calculated using Segment-
taion Metrics Library (Jia, 2020).
ROBOVIS 2021 - 2nd International Conference on Robotics, Computer Vision and Intelligent Systems
124
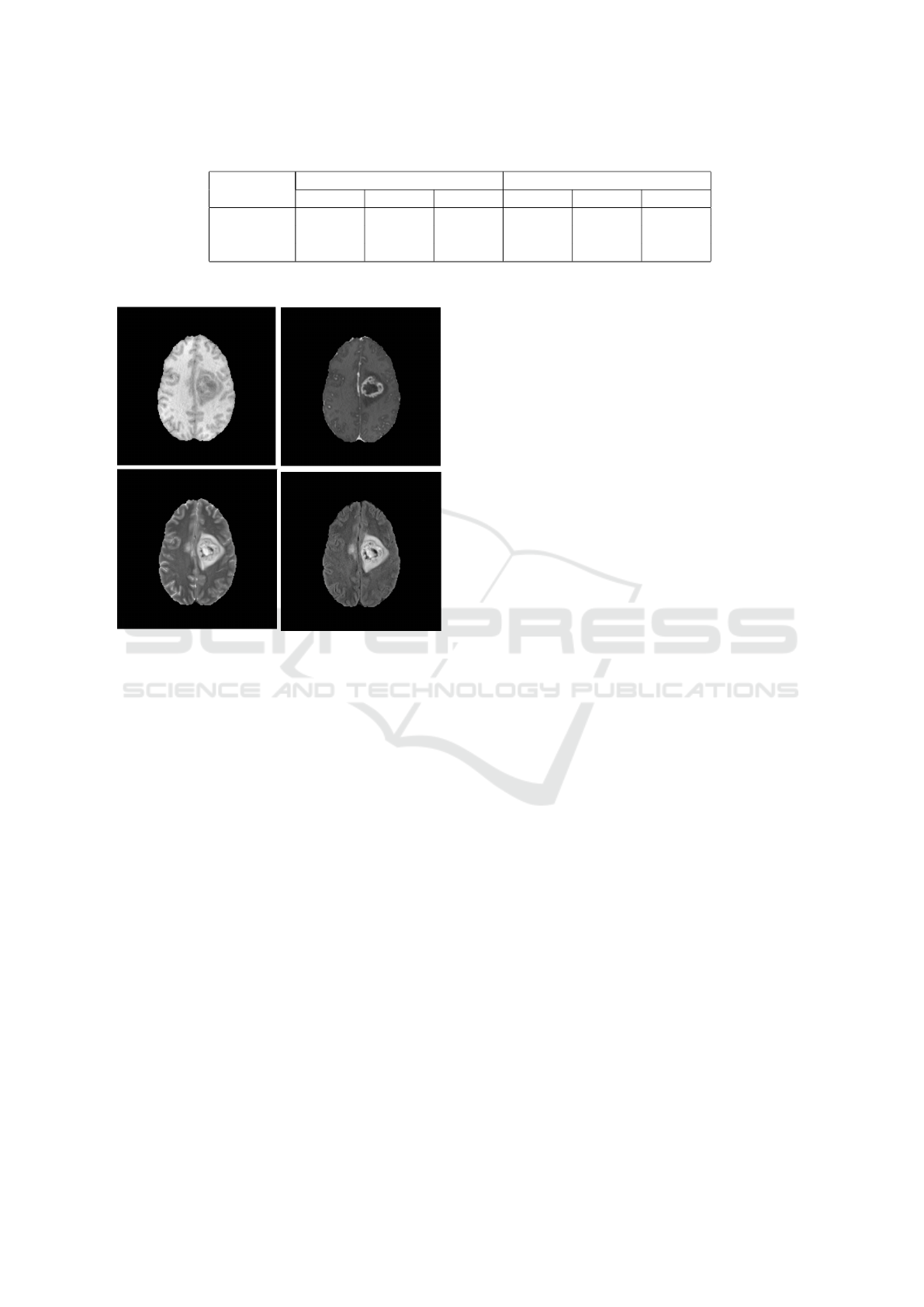
Table 1: Experiment results. The result is calculated based on new training data in BRATS2020 dataset.
Model
Dice Score Hausdorff95
ET TC WT ET TC WT
UNet 0.82585 0.84112 0.91564 2.68228 7.98642 7.48138
CAUNet 0.80907 0.83106 0.91346 5.90530 8.29032 7.86522
MCAUNet 0.82006 0.84607 0.91898 5.44802 7.55089 7.26619
Figure 4: Four modalities of an MRI scan layer. On the first
row is T1 Image and T1ce Image. The second row contain
T2 Image and Flair Image.
4.3 Model Comparison
We can qualitatively compare the difference between
each model in Figure 5. As we can see MCA-UNet
and CA-UNet have better segment for necrotic and
non-enhancing tumor core (NCR/NET) and. Further
more MCA-UNet’s segmentation of NCR/NET have
better structure than CA-UNet.
For more detail comparing our model, we calcu-
late Dice Score, and 95 percentile Hausdorff Distance
for three segment target: Whole Tumor (WT), Tumor
Core (TC) and Enhance Tumor (ET). Table 1 record
result of both score.
As we can see, UNet still shine above all model
in segmenting Enhance Tumor. But in the rest targets,
MCA-UNet scored higher than the orginal UNet. Fur-
thermore, CA-UNet scored lowest among them. But
with the modification in MCA-UNet, the scored im-
proved significantly, which proved the effectiveness
of our proposal.
5 DISCUSSION AND
CONCLUSION
In this section, We describe what we haven’t done in
this article and how we can improve the performance
of our work. The first topic is Data Augmentation, the
second topic is moving to 3D model
5.1 Data Augmentation
Data imbalance and intensity normalization are two
biggest issue MRI scans. In this section we discuss
and how to handle those two issue and also how we
made used of the four modalities of one MRI scans.
As we can see in Figure 5. The tumor region
is only cover around 10% of the whole image and
around 30% of the brain, which cause the imbalance
between segment mask and the image. To handle this
issue, we can crop the image to make the brain and
segment mask cover more percent of the image. In
our work, we did not cropped the images because we
want to evaluate the model efficient without data aug-
mentation.
To handle data normalization, an effective method
is applying N4 Bias Correction (Tustison et al., 2010).
The N4 bias field correction is a famous algorithm to
handle the low frequency bias in MRI scans which
make the intensity become uniform. In future work,
we can apply this algorithm to do more experiment.
MRI scans is a 3D data which is not fitted for our
model. Therefore we decided to slice each layer of the
MRI scans and used it as the input of our model. This
method prevent us from using the relationship of z-
axis feature of each image. Instead of the z-axis rela-
tionship, we make use of four modalities of the image
by compressing four modalities in to one 2D image
with four channels. To make use of this setup is one
of the reason why we chose the coordinate attention
module which have ability to enhancing channel-wise
relationships.
5.2 Moving to 3D
As we mentioned in section 5.1, our work focus on
2D model. This method removed the z-axis relation-
Coordinate Attention UNet
125
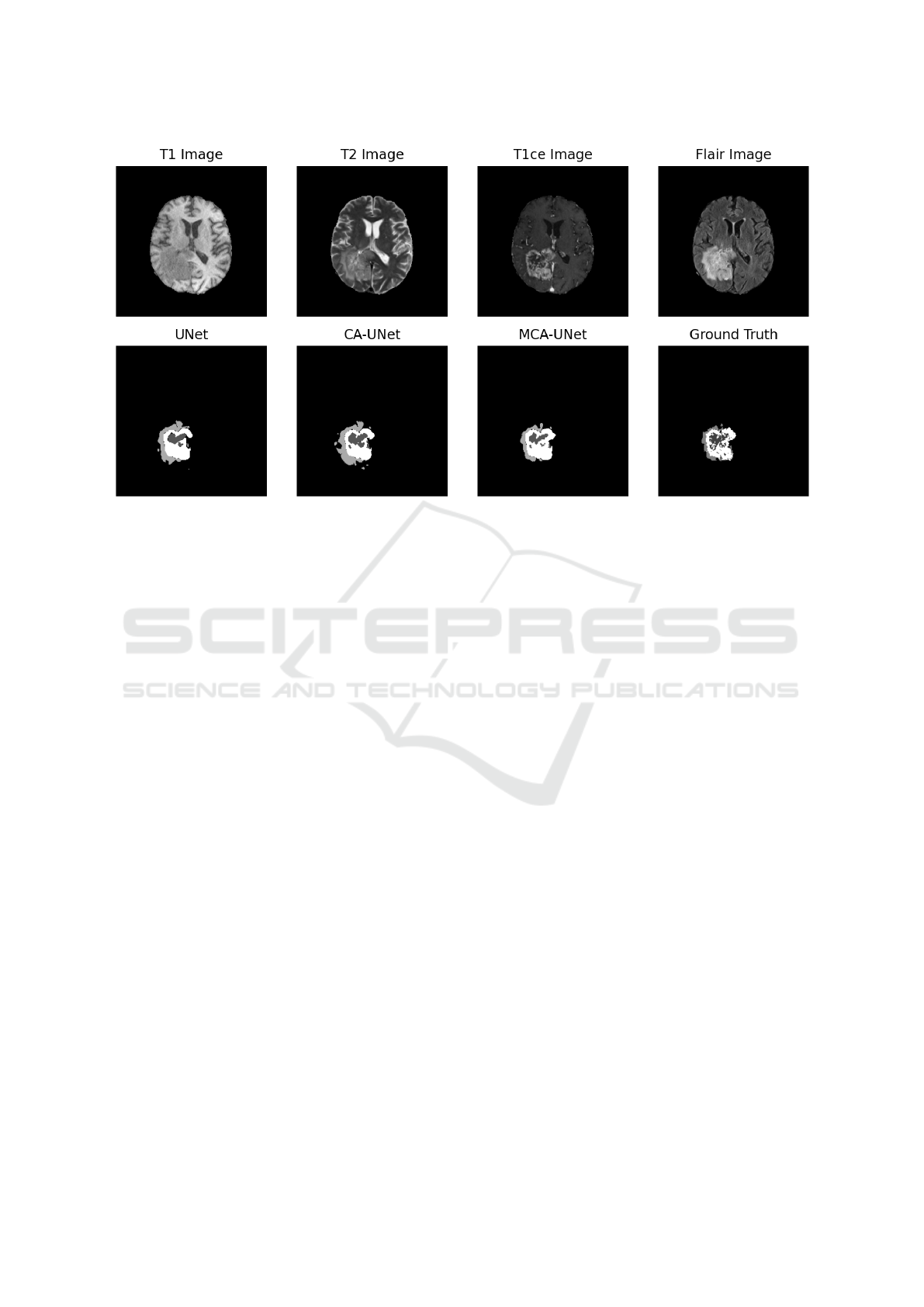
Figure 5: Segmentation result of three models. The first row is the four image types of the input image. The second row show
the prediction of UNet, Coordinate Attention UNet (CA-UNet), Max pooling Coordinate Attention UNet (MCA-UNet) and
Ground Truth. GD-enhancing tumor (ET) is colored white, the peritumoral edema (ED) is colored light grey, and the necrotic
and non-enhancing tumor core (NCR/NET) is colored dark grey.
ship, which is an important feature of 3D data. There-
fore in the futures, we can continue our research with
applying attention to 3D model to encode the z-axis
relationship.
In this research, we also found that compressing
all modalities into one image have positive affect on
the results of our model. In the future work, we can
exploring the effectiveness of this method with 3D
data.
5.3 Conclusion
In this paper, we proposed a new UNet model, which
utilize the attention mechanism to solve the con-
text loss and feature dilution of original UNet. We
also proposed a modification of Coordinate Attention
Module to be more suitable with the MRI scans data.
These proposal have been proved effective with our
experiments.
REFERENCES
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M.,
Crimi, A., Shinohara, R. T., Berger, C., Ha, S. M.,
Rozycki, M., Prastawa, M., Alberts, E., Lipkov
´
a, J.,
Freymann, J. B., Kirby, J. S., Bilello, M., Fathallah-
Shaykh, H. M., Wiest, R., Kirschke, J., Wiestler, B.,
Colen, R. R., Kotrotsou, A., LaMontagne, P., Marcus,
D. S., Milchenko, M., Nazeri, A., Weber, M., Ma-
hajan, A., Baid, U., Kwon, D., Agarwal, M., Alam,
M., Albiol, A., Albiol, A., Varghese, A., Tuan, T. A.,
Arbel, T., Avery, A., B., P., Banerjee, S., Batchelder,
T., Batmanghelich, K. N., Battistella, E., Bendszus,
M., Benson, E., Bernal, J., Biros, G., Cabezas, M.,
Chandra, S., Chang, Y., and et al. (2018). Identify-
ing the best machine learning algorithms for brain tu-
mor segmentation, progression assessment, and over-
all survival prediction in the BRATS challenge. CoRR,
abs/1811.02629.
Boureau, Y.-L., Bach, F., LeCun, Y., and Ponce, J. (2010).
Learning mid-level features for recognition. In 2010
IEEE Computer Society Conference on Computer Vi-
sion and Pattern Recognition. IEEE.
Chen, C., Liu, X., Ding, M., Zheng, J., and Li, J. (2019). 3d
dilated multi-fiber network for real-time brain tumor
segmentation in MRI. CoRR, abs/1904.03355.
C¸ ic¸ek,
¨
O., Abdulkadir, A., Lienkamp, S. S., Brox, T.,
and Ronneberger, O. (2016). 3d u-net: Learning
dense volumetric segmentation from sparse annota-
tion. CoRR, abs/1606.06650.
Havaei, M., Davy, A., Warde-Farley, D., Biard, A.,
Courville, A. C., Bengio, Y., Pal, C., Jodoin, P., and
Larochelle, H. (2015). Brain tumor segmentation with
deep neural networks. CoRR, abs/1505.03540.
Hou, Q., Zhou, D., and Feng, J. (2021). Coordinate atten-
tion for efficient mobile network design. In CVPR.
Hu, J., Shen, L., and Sun, G. (2017). Squeeze-and-
excitation networks. CoRR, abs/1709.01507.
Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., Iwamoto,
Y., Han, X., Chen, Y.-W., and Wu, J. (2020). Unet
3+: A full-scale connected unet for medical image
segmentation.
ROBOVIS 2021 - 2nd International Conference on Robotics, Computer Vision and Intelligent Systems
126

Isensee, F., Petersen, J., Klein, A., Zimmerer, D., Jaeger,
P. F., Kohl, S., Wasserthal, J., Koehler, G., Norajitra,
T., Wirkert, S. J., and Maier-Hein, K. H. (2018). nnu-
net: Self-adapting framework for u-net-based medical
image segmentation. CoRR, abs/1809.10486.
Jia, J. (2020). A package to compute segmentation metrics:
seg-metrics.
Lou, A., Guan, S., and Loew, M. H. (2021). DC-UNet: re-
thinking the u-net architecture with dual channel effi-
cient CNN for medical image segmentation. In Land-
man, B. A. and I
ˇ
sgum, I., editors, Medical Imaging
2021: Image Processing. SPIE.
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J.,
Farahani, K., Kirby, J., Burren, Y., Porz, N., Slot-
boom, J., Wiest, R., Lanczi, L., Gerstner, E., We-
ber, M.-A., Arbel, T., Avants, B. B., Ayache, N.,
Buendia, P., Collins, D. L., Cordier, N., Corso, J. J.,
Criminisi, A., Das, T., Delingette, H., Demiralp, C.,
Durst, C. R., Dojat, M., Doyle, S., Festa, J., Forbes,
F., Geremia, E., Glocker, B., Golland, P., Guo, X.,
Hamamci, A., Iftekharuddin, K. M., Jena, R., John,
N. M., Konukoglu, E., Lashkari, D., Mariz, J. A.,
Meier, R., Pereira, S., Precup, D., Price, S. J., Ra-
viv, T. R., Reza, S. M. S., Ryan, M., Sarikaya, D.,
Schwartz, L., Shin, H.-C., Shotton, J., Silva, C. A.,
Sousa, N., Subbanna, N. K., Szekely, G., Taylor, T. J.,
Thomas, O. M., Tustison, N. J., Unal, G., Vasseur, F.,
Wintermark, M., Ye, D. H., Zhao, L., Zhao, B., Zi-
kic, D., Prastawa, M., Reyes, M., and Leemput, K. V.
(2015). The multimodal brain tumor image segmen-
tation benchmark (BRATS). IEEE Transactions on
Medical Imaging, 34(10):1993–2024.
Milletari, F., Navab, N., and Ahmadi, S. (2016). V-net:
Fully convolutional neural networks for volumetric
medical image segmentation. CoRR, abs/1606.04797.
Myronenko, A. (2018). 3d MRI brain tumor segmen-
tation using autoencoder regularization. CoRR,
abs/1810.11654.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M. C. H., Hein-
rich, M. P., Misawa, K., Mori, K., McDonagh, S. G.,
Hammerla, N. Y., Kainz, B., Glocker, B., and Rueck-
ert, D. (2018). Attention u-net: Learning where to
look for the pancreas. CoRR, abs/1804.03999.
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J.,
Chanan, G., Killeen, T., Lin, Z., Gimelshein, N.,
Antiga, L., Desmaison, A., K
¨
opf, A., Yang, E., De-
Vito, Z., Raison, M., Tejani, A., Chilamkurthy, S.,
Steiner, B., Fang, L., Bai, J., and Chintala, S. (2019).
Pytorch: An imperative style, high-performance deep
learning library. CoRR, abs/1912.01703.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. CoRR, abs/1505.04597.
Sinha, A. and Dolz, J. (2019). Multi-scale guided at-
tention for medical image segmentation. CoRR,
abs/1906.02849.
Tamimi, A. F. and Juweid, M. (2017). Epidemiology and
outcome of glioblastoma. In Glioblastoma, pages
143–153. Codon Publications.
Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan,
A., Yushkevich, P. A., and Gee, J. C. (2010). N4itk:
Improved n3 bias correction. IEEE Transactions on
Medical Imaging, 29(6):1310–1320.
Zhou, C., Ding, C., Wang, X., Lu, Z., and Tao, D.
(2019). One-pass multi-task networks with cross-task
guided attention for brain tumor segmentation. CoRR,
abs/1906.01796.
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang,
J. (2018). Unet++: A nested u-net architecture for
medical image segmentation. CoRR, abs/1807.10165.
Coordinate Attention UNet
127
