
Evo-Path: Querying Data Evolution through Complex Changes
Theodora Galani
1
, Yannis Stavrakas
2
, George Papastefanatos
2
and Yannis Vassiliou
1
1
School of Electrical and Computer Engineering, NTUA, Iroon Polytechniou 9, Athens, Greece
2
RC ATHENA, Artemidos 6 & Epidavrou, Marousi, Greece
Keywords: Querying Data Evolution, Change Modelling, XPath.
Abstract: Evo-graph is a model for data evolution that captures data versions and treats changes as first-class citizens.
A change in evo-graph can be compound, comprising disparate changes, and is associated with the data
items it affects. In previous work, we specified how an evo-graph can be reduced to a snapshot holding
under a specific time instance, we presented an XML representation of evo-graph called evoXML, we
defined how evo-graph is constructed as the current snapshot evolves, as well as presented and evaluated the
C2D framework that implements these concepts using XML technologies. In this paper, we formally define
evo-path, an XPath extension for querying the data history and change structure in a uniform way over evo-
graph. We specify the evo-path syntax, semantics and implementation, and present several query categories.
1 INTRODUCTION
The dynamic nature of web data poses new
challenges for data management. In particular,
revisiting past data snapshots may not be enough for
users of scientific data. Additionally, they would like
to review how and why data have evolved, in order
to reassess and compare previous and current results.
Such an activity may require a search that moves
backwards and forwards in time, spread across
disparate parts of a database, and perform complex
queries on the semantics of the changes that
modified the data. The need for tracing past changes
and data lineage is evident in a wide range of web
information management domains.
Consider an example taken from Biology, the
revision in the classification of diabetes, which was
caused by a better understanding of insulin (National
research council, 2005). Initially, diabetes was
classified according to the age of the patient, as
juvenile or adult onset. As the role of insulin became
clearer two more subcategories were added: insulin
dependent and non-insulin dependent. All juvenile
cases of diabetes are insulin dependent, while adult
onset may be either insulin dependent or non-insulin
dependent. In Figure 1, the leftmost/rightmost image
depicts a tree representation of the initial/revised
diabetes classification. Supposing that a scientist
examines the revised classification, she may realize
that diabetes categories are not as expected. She
would like to know: Which may be the previous
structure of categories? Which changes are
responsible for the reorganization of diabetes
categories? What are the previous versions of the
data nodes that changed due to the reorganization of
diabetes categories? However, these representations
are not informative on which parts of the data
evolved and how, which changes led from one
version to another, or what changes were applied on
which parts of data. Recording change operations in
a log or computing deltas between successive
versions do not solve the problem. In most cases, it
is difficult to interpret a posteriori isolated
operations because they usually form more complex,
semantically rich changes, each comprising many
small changes on disparate parts of data. As a result,
answering such questions may require complex
queries in different parts of a database, a task which
may be even more intensive for large datasets.
We argue that in systems where evolution issues
are paramount, changes should not be treated solely
as transformation operations on the data, but rather
as first class citizens retaining structural, semantic,
and temporal characteristics. In previous work, we
proposed a graph model, evo-graph (Stavrakas and
Papastefanatos, 2010), and its XML representation,
evoXML (Stavrakas and Papastefanatos, 2011),
capturing relationships between evolving data and
changes applied on them. Evo-graph models changes
explicitly as first class citizens and thus, enables
Galani, T., Stavrakas, Y., Papastefanatos, G. and Vassiliou, Y.
Evo-Path: Quer ying Data Evolution through Complex Changes.
DOI: 10.5220/0010615703530361
In Proceedings of the 10th International Conference on Data Science, Technology and Applications (DATA 2021), pages 353-361
ISBN: 978-989-758-521-0
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
353
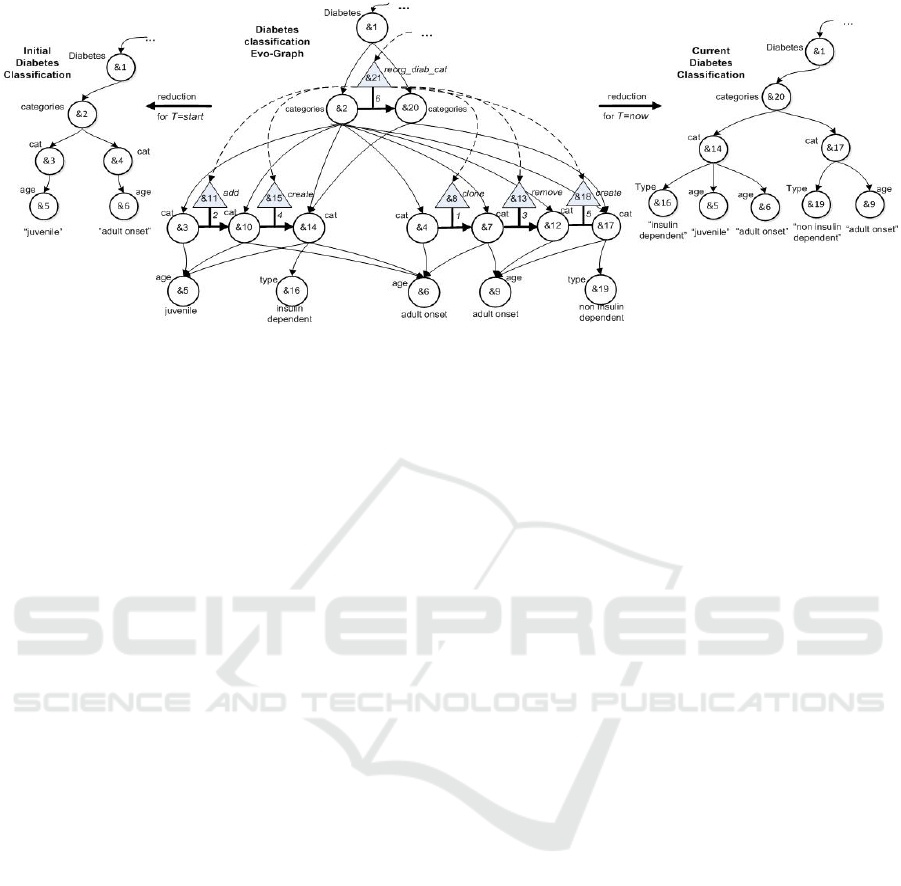
Figure 1: Snap-models of diabetes classification before (left) and after (right) revision and the relevant evo-graph (middle).
querying data and changes in a uniform way. In
(Papastefanatos et al., 2013) we showed how evo-
graph is constructed, recording data history and
structured changes step by step as current snapshot
evolves, and evaluated the C2D framework, which
implements these concepts via XML technologies.
In this paper, we formally define evo-path, an
XPath (Robie, Dyck and Spiegel, 2017) extension
for performing time- and change-aware queries on
evo-graph. Evo-path allows querying both data
history and change structure in a uniform way,
taking advantage of changes in order to retrieve and
relate data that are otherwise irrelevant to each other.
Temporal, evolution and causality queries are
supported. In 0(Stavrakas and Papastefanatos, 2010)
and (Stavrakas and Papastefanatos, 2011) evo-path
was introduced, but only a syntax outline and a by-
example translation into equivalent XQuery
expressions over evoXML were presented. In this
paper, we contribute the following: a) we enrich the
evo-path syntax, b) we define evo-path formal
semantics, c) we present an implementation based
on a formal translation of evo-path into equivalent
XPath expressions over evoXML.
Section 2 covers previous work on evo-graph,
evoXML, basic and complex changes. Section 3
formally defines evo-path: syntax, semantics,
implementation and examples. Section 4 revises
related work. Section 5 concludes the paper.
2 PRELIMINARIES
Snap-model. In terms of this work, we assume that
data is represented by a rooted, node-labelled, leaf-
valued tree called snap-model. A snap-model S(V,
E) consists of a set of nodes V, divided into complex
and atomic, with atomic being the tree leaves, and a
set of directed edges E. At any time instance, snap-
model undergoes arbitrary changes.
Evo-graph. An evo-graph G is a graph-based
model that captures all the instances of an evolving
snap-model across time, together with the changes
responsible for the transitions. It consists of: Data
nodes (complex and atomic) and data edges,
departing from every complex data node. Change
nodes (complex and atomic), representing change
events, depicted as triangles to distinguish from
circular data nodes. Change edges connect every
complex change node to change nodes it contains.
Evolution edges connect each change node with two
data nodes, the version before and after the change.
r
D
is the data root, such that there is a path formed
by data edges from r
D
to every other data node. r
C
is
the change root, such that there is a path formed by
change edges from r
C
to every other change node.
Intuitively, evo-graph consists of a data graph,
holding the data versions, and a tree of changes,
which interconnect via evolution edges. Change
nodes are annotated with timestamps denoting the
time instance each change occurred. Although valid
time may be considered, we rely on transaction time,
assuming a linear time domain constituted by
consecutive discrete values and two special time
instances: 0 for the beginning of time and now for
the current time. Timestamps are used for
determining the validity timespan of data nodes and
data edges in evo-graph. Evo-graph can then be
reduced to a snap-model holding under a specified
time instance through the reduction process
(Stavrakas and Papastefanatos 2010).
An evo-graph example is the middle image in
Figure 1, representing the revision in the diabetes
classification from the graph of Figure 1 left to right.
The revision process is denoted by the complex
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
354

change reorg_diab_cat (node &21) composed by 5
basic changes (in order they occurred): clone (node
&8), add (node &11), remove (node &13), create
(node &15), and create (node &18). The reduction
of the evo-graph for T=start (i.e. 0)/now results in
the snap-model of the leftmost/rightmost image of
Figure 1.
EvoXML. In (Stavrakas and Papastefanatos,
2011) we presented an XML representation of evo-
graph, the evoXML. Table 1 presents the evoXML
for time instance 1 of the evo-graph in Figure 1,
including only the clone operation (node &8, lines
12-16, 19-20). Notice that the edge from node &7 to
node &6 (denoting that &6 remains a child of the
next version of &4) is represented via the evoXML
reference evo:ref in line 14, which points to the
element in line 10. Also, notice the change node &8
in lines 19-20. Comparing to (Stavrakas and
Papastefanatos 2011), we explicitly encode the
timespan of each data node on it, via attributes
evo:ts and evo:te, to facilitate evo-path evaluation.
Basic and Complex Changes. The following
basic change operations may be applied on a snap-
model: create, add, remove, update, clone. A
complex change applied on a node of a snap-model
is a sequence of basic and other complex change
operations that are applied on the node itself or/and
its descendants, formulating semantically coherent
sequences (Papastefanatos et al., 2013). A complex
change example is reorg_diab_cat applied on
categories node of Figure's 1 leftmost image:
reorg-diab-cat(&2) { clone(&4, &6, &9)
add(&3, &6) remove(&4, &6) create(&3, &16,
'type', 'insulin dependent') create(&4, &19,
'type', 'non insulin dependent') }
Table 1: EvoXML for time instance 1.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
<evo:evoXML xmlns=”” xmlns:evo=”http://web.
imis.athena-innovation.gr/projects/c2d”>
<evo:DataRoot evo:id=”dataroot”>
<Diabetes evo:id=”1” evo:ts="0" evo:te="now">
<categories evo:id=”2” evo:ts="0" evo:te="now">
<cat evo:id=”3”evo:ts="0" evo:te="now">
<age evo:id=”5”evo:ts="0" evo:te="now">
juvenile</age></cat>
<cat evo:id=”4”evo:ts="0" evo:te="1">
<age evo:id=”6”evo:ts="0" evo:te="now">
adult onset</age></cat>
<cat evo:id=”7” evo:ts=”1” evo:te="now"
evo:previous=”4”>
<age evo:ref=”6”/>
<age evo:id=”9”evo:ts=”1” evo:te="now">
adult onset</age></cat>
</categories></Diabetes></evo:DataRoot>
<evo:ChangeRoot evo:id=”changeroot”>
<clone evo:id=”8” evo:tt=”1” evo:before=”4”
evo:after=”7”/></evo:ChangeRoot>
</evo:evoXML >
3 EVO-PATH
3.1 Syntax
Similar to XPath, evo-path uses path expressions to
move through and select data nodes. In addition,
evo-path allows the navigation through change
nodes on evo-graph. Consequently, there are two
types of path expressions in evo-path: data path and
change path expressions. Also, several predicates
are supported to express conditions on evo-graph
temporal properties and evolution edges.
Data path expressions start from the data root of
evo-graph and return data nodes. Similar to XPath,
they are written as a sequence of location steps
separated by “/” characters and shortcuts can be used
as in the two equivalent evo-paths below:
/child::A/
descendant-or-self::node()/
child::B/child::*[position()=1]
/A//B/*[1]
Change path expressions start from the change
root of evo-graph and return change nodes. They
have the same syntax as data path expressions, but
are enclosed in square brackets:
</location_step1/…/location_stepN>
Temporal predicates are introduced in evo-path
in order to express temporal conditions on the evo-
graph nodes. The following types are employed:
1) On data node timespan:
[ts() operator (t_1, t_2)], where ts()
evaluates to the validity timespan of the context data
node,
operator may be [not] (in |
contains | meets | equals) covering the
standard operations between sets, allowing the use
of
not in front of any of the operators, and t_1,
t_2
are specified timestamps defining a timespan.
[ts() operator t], where ts() evaluates to
the validity timespan of the context data node,
operator may be [not] covers, and t is a
specified timestamp, for the case where a specified
timestamp exists or not in the validity timespan.
2) On data node timespan start time:
[tstart() operator t], where tstart()
evaluates to the start of the validity timespan of the
context data node,
operator may be (> | >= | =
| < | <=), and t is a specified timestamp.
3) On data node timespan end time:
[tend() operator t], where tend()
evaluates to the end of the validity timespan of the
Evo-Path: Querying Data Evolution through Complex Changes
355

context data node (operator and t as in case 2).
4) On change node timestamp:
[tt() operator t], where tt() evaluates to
the timestamp of the context change node
(
operator and t as in case 2).
Evolution predicates are used to assert the
existence of evolution edges at specific points in the
graph. They combine a data path expression with a
change path expression and vice versa, implying that
the specified data are affected by the specified
change. Their general form is:
5)
data_path_expr
[evo-filter(<change_path_expr>)]
6) <change_path_expr
[evo-filter(data_path_expr)]>
where evo-filter may be one of: evo-
before(), evo-after() and evo-both().
Each
evo-filter evaluates into true or false, in
case there is or not an evolution edge involving the
data or change node in context.
evo-before() and
evo-after() refer on a specific side of the
evolution edge, while
evo-both()on both sides. In
case 5
evo-before() and evo-after() validate
whether the data node in context holds before and
after respectively the application of the change being
represented by the change node in context.
evo-
both() validates whether the data node holds
either before or after the change. In case 6
evo-
before() and evo-after() validate whether the
change node in context represents the change before
and after which the data node in context holds
respectively.
evo-both() validates whether the
change node represents the change either before or
after which the data node holds.
3.2 Example Queries
The evo-path examples refer to and are evaluated on
the evo-graph of Figure 1 regarding diabetes.
1) Temporal queries - Querying the history of
data elements: Suppose that a scientist examines the
current diabetes snapshot and realizes that the
categories structure is not as expected. She wants to
retrieve the previous versions of data node &20.
//Diabetes/categories [ts() not covers
'now'] (Q1)
This is a data path expression with a temporal
predicate that evaluates false for the current version
of
categories and true for every other version. It
returns node &2 with timespan [0, 5].
2) Evolution queries - Querying changes applied
on data elements: The scientist observes the creation
of several new nodes under the categories node. She
wants to know the complex changes that contain a
relevant create operation, to check if create was part
of a larger modification.
<//* [evo-both(//Diabetes//*)]
[.//create [evo-both(//Diabetes/
categories/cat)]]> (Q2)
This is a change path expression. The first
predicate is an evolution predicate for returning all
the change nodes that are applied to
Diabetes node
or any of its descendants. The second predicate
dictates that only changes with a
create descendant
applied on a
cat object can be returned. It returns
node &21 with timestamp 6, i.e. the complex change
reorg_diab_cat, affecting data node &2 and
resulting into data node &20.
The scientist can now retrieve all the changes
associated with
reorg_diab_cat, in order to
understand its full effect.
<//reorg_diab_cat/*> (Q3)
This change path expression returns nodes &8,
&11, &13, &15 and &18.
3) Causality queries - Querying relationships
between change and data elements: Realizing that
the modifications on diabetes categories are related
to the complex change &21
reorg_diab_cat, the
scientist decides to check all the previous versions of
the data nodes affected by
reorg_diab_cat and its
descendant changes.
//* [evo-before(
<//reorg_diab_cat//*>)] (Q4)
The data path expression returns all data nodes
being connected through evolution edges with a
reorg_diab_cat change node (&21) or one of its
descendant change nodes, specifically those before
each change due to
evo-before(). The nodes &3
with timespan [0, 1], &4 [0, 1), &7 [1, 2], &10 [2, 3]
and &12 [3, 4] are returned. The scientist now
realizes the sequence of data evolution.
3.3 Semantics
In XPath, the meaning of a path expression is the
sequence of nodes, at the end of each path, that
matches the expression. In evo-path, the meaning of
a data path expression is a sequence of (data-node,
interval) pairs such that the data-node has been at the
end of a matching data path continuously during that
interval. The interval is the validity timespan of the
data-node. In evo-path, the meaning of a change
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
356

path expression is a sequence of (change-node,
instance, data-node-before, data-node-after) tuples
such that the change-node is at the end of a matching
change path at the specific instance and it references
the data-node-before and the data-node-after the
change. The instance is the timestamp (transaction
time) when the change was applied on the data-
node-before, leading to the data-node-after.
For specifying the evo-path semantics the formal
XPath semantics introduced by (Wadler, 1999) have
been adapted. The meaning of an XPath expression
is specified with respect to a context node. For a data
path expression, this is extended to a context pair of
a data-node and a time interval. For a change path
expression, its meaning is specified with respect to a
context tuple of a change-node, a time instance, a
data-node before and data-node after the change. For
the data part, four semantic functions are defined:
𝑆,𝑄,𝑄
and 𝑄
. 𝑆
⟦
𝑝
⟧
𝑥 denotes the sequence of pairs
(data-node, interval) selected by pattern 𝑝 when 𝑥 is
the context pair. It may also denote a sequence of
values. The boolean expression 𝑄
⟦
𝑞
⟧
𝑥 denotes
whether or not the qualifier 𝑞 is satisfied when the
context pair (data-node, interval) is 𝑥. The boolean
expression 𝑄
⟦
𝑞
⟧
𝑥 denotes whether or not a
temporal condition 𝑞
is satisfied, while the boolean
Table 2: Formal Semantics of Evo-Path.
𝑆
⟦
/𝑝
⟧
𝑥𝑆
⟦
𝑝
⟧
𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡
𝑥
;
𝑆
⟦
//𝑝
⟧
𝑥 𝑥
𝑥
∈ 𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡
𝑥
, 𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑆
⟦
𝑝
/𝑝
⟧
𝑥
𝑣
,𝐼
∩𝐼
|
𝑣
,𝐼
∈𝑆
⟦
𝑝
⟧
𝑥,
𝑣
,𝐼
∈𝑆
⟦
𝑝
⟧
𝑣
,𝐼
;
𝑆
⟦
𝑝
//𝑝
⟧
𝑥
𝑥
|
𝑥
∈𝑆
⟦
𝑝
⟧
𝑥,𝑥
∈𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠
𝑥
,𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑆
⟦
𝑝
𝑞
⟧
𝑥
𝑣,𝐼
|
𝑣,𝐼
∈𝑆
⟦
𝑝
⟧
𝑥,𝑄
⟦
𝑞
⟧
𝑣,𝐼
;
𝑆
⟦
𝑛
⟧
𝑥
𝑣,𝐼
|
𝑖𝑠𝐸𝑙𝑒𝑚𝑒𝑛𝑡
𝑣
,𝑐ℎ𝑖𝑙𝑑
𝑥
𝑣,𝐼
,𝑛𝑎𝑚𝑒
𝑣
𝑛
;
𝑆
⟦
𝑡𝑠𝑡𝑎𝑟𝑡
⟧
𝑥
𝑠
|
𝑥
𝑣,𝐼
,𝐼
𝑠,𝑒
;
𝑆
⟦
𝑡𝑒𝑛𝑑
⟧
𝑥
𝑒
|
𝑥
𝑣,𝐼
,𝐼
𝑠, 𝑒
;
𝑆
⟦
𝑝
𝑞
⟧
𝑥
𝑣,𝐼
|
𝑣,𝐼
∈𝑆
⟦
𝑝
⟧
𝑥,𝑄
⟦
𝑞
⟧
𝑣,𝐼
;
𝑆
⟦
𝑎𝑛𝑐𝑒𝑠𝑡𝑜𝑟 ∶∶ 𝑝
⟧
𝑥
𝑥
|
𝑥
∈𝑝𝑟𝑒𝑛𝑜𝑑𝑒𝑠
𝑥
,𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑄
⟦
𝑝𝑠
⟧
𝑥
𝑣,𝐼
|
𝑣,𝐼
∈𝑆
⟦
𝑝
⟧
𝑥,𝑣𝑎𝑙𝑢𝑒
𝑣
𝑠
∅;
𝑄
⟦
𝑝
⟧
𝑥
𝑥
|
𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
∅;
𝑄
⟦
𝑡𝑠
𝑖𝑛
𝑡
,𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡
𝑡
,𝑡
𝑡
∅;
𝑄
⟦
𝑡𝑠
𝑐𝑜𝑛𝑡𝑎𝑖𝑛𝑠
𝑡
,𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡
𝑡
,𝑡
𝑡
∅;
𝑄
⟦
𝑡𝑠
𝑚𝑒𝑒𝑡𝑠
𝑡
,𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,
𝑡
,𝑡
∩
𝑡
,𝑡
∅
∅;
𝑄
⟦
𝑡𝑠
𝑒𝑞𝑢𝑎𝑙𝑠
𝑡
,𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡
𝑡
,𝑡
𝑡
∅;
𝑄
⟦
𝑡𝑠
𝑐𝑜𝑣𝑒𝑟𝑠 𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡𝑡
,𝑡𝑡
∅;
𝑄
⟦
𝑡𝑠𝑡𝑎𝑟𝑡
𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡
𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
∅;
𝑄
⟦
𝑡𝑒𝑛𝑑
𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣,
𝑡
,𝑡
,𝑡
𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑏𝑒𝑓𝑜𝑟𝑒
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑥
𝑥
|
𝑥
𝑣,𝐼
,
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑟
,𝑣𝑣
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑎𝑓𝑡𝑒𝑟
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑥
𝑥
|
𝑥
𝑣,𝐼
,
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑟
,𝑣𝑣
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑏𝑜𝑡ℎ
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑥
𝑥
|
𝑥
𝑣,𝐼
,
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
〈
𝑐ℎ𝑎𝑛𝑔𝑒_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
〉
⟧
𝑟
,𝑣𝑣
∨ 𝑣 𝑣
∅;
𝑆
⟦
〈
/𝑝
〉
⟧
𝑥𝑆
⟦
𝑝
⟧
𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡
𝑥
;
𝑆
⟦
〈
//𝑝
〉
⟧
𝑥 𝑥
𝑥
∈ 𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠
𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡
𝑥
, 𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑆
⟦
〈
𝑝
/𝑝
〉
⟧
𝑥
𝑥
|
𝑥
∈𝑆
⟦
𝑝
⟧
𝑥,𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑆
⟦
〈
𝑝
//𝑝
〉
⟧
𝑥
𝑥
|
𝑥
∈𝑆
⟦
𝑝
⟧
𝑥,𝑥
∈ 𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠
𝑥
,𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑆
⟦
〈
𝑝
𝑞
〉
⟧
𝑥
𝑣
,𝑖,𝑣
,𝑣
|
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
𝑝
⟧
𝑥,𝑄
⟦
𝑞
⟧
𝑣
,𝑖,𝑣
,𝑣
;
𝑆
⟦
𝑛
⟧
𝑥
𝑣
,𝑖,𝑣
,𝑣
|
𝑖𝑠𝐸𝑙𝑒𝑚𝑒𝑛𝑡
𝑣
,𝑐ℎ𝑖𝑙𝑑
𝑥
𝑣
,𝑖,𝑣
,𝑣
,𝑛𝑎𝑚𝑒
𝑣
𝑛
;
𝑆
⟦
𝑡𝑡
⟧
𝑥
𝑖
|
𝑥
𝑣
,𝑖,𝑣
,𝑣
;
𝑆
⟦
〈
𝑝
𝑞
〉
⟧
𝑥
𝑣
,𝑖,𝑣
,𝑣
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
𝑝
⟧
𝑥,𝑄
⟦
𝑞
⟧
𝑣
,𝑖,𝑣
,𝑣
;
𝑆
⟦
〈
𝑎𝑛𝑐𝑒𝑠𝑡𝑜𝑟 ∶∶ 𝑝
〉
⟧
𝑥
𝑥
|
𝑥
∈𝑝𝑟𝑒𝑛𝑜𝑑𝑒𝑠
𝑥
,𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
;
𝑄
⟦
𝑝𝑠
⟧
𝑥
𝑣
,𝑖,𝑣
,𝑣
|
𝑣
,𝑖,𝑣
,𝑣
∈𝑆
⟦
𝑝
⟧
𝑥,𝑣𝑎𝑙𝑢𝑒
𝑣
𝑠
∅;
𝑄
⟦
𝑝
⟧
𝑥
𝑥
|
𝑥
∈𝑆
⟦
𝑝
⟧
𝑥
∅;
𝑄
⟦
𝑡𝑡
𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
⟧
𝑥
𝑥
|
𝑥
𝑣
,𝑖,𝑣
,𝑣
,𝑖 𝑜𝑝𝑒𝑟𝑎𝑡𝑜𝑟 𝑡
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑏𝑒𝑓𝑜𝑟𝑒
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑥
𝑥
|
𝑥
𝑣
,𝑖,𝑣
,𝑣
,
𝑣,𝐼
∈𝑆
⟦
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑟
,𝑣𝑣
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑎𝑓𝑡𝑒𝑟
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑥
𝑥
|
𝑥
𝑣
,𝑖,𝑣
,𝑣
,
𝑣,𝐼
∈𝑆
⟦
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑟
,𝑣𝑣
∅;
𝑄
⟦
𝑒𝑣𝑜 𝑏𝑜𝑡ℎ
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑥
𝑥
|
𝑥
𝑣
,𝑖,𝑣
,𝑣
,
𝑣,𝐼
∈𝑆
⟦
𝑑𝑎𝑡𝑎_𝑝𝑎𝑡ℎ_𝑒𝑥𝑝𝑟
⟧
𝑟
,𝑣𝑣
∨ 𝑣 𝑣
∅;
Where: 𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠
𝑦
𝑣,𝐼
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑑𝑎𝑡𝑎 𝑝𝑎𝑡ℎ 𝑓𝑟𝑜𝑚 𝑦 𝑡𝑜 𝑣 𝑎𝑛𝑑 𝐼 𝑖𝑠 𝑡ℎ𝑒 𝑣𝑎𝑙𝑖𝑑𝑖𝑡𝑦 𝑡𝑖𝑚𝑒𝑠𝑝𝑎𝑛 𝑜𝑓 𝑣
𝑝𝑟𝑒𝑛𝑜𝑑𝑒𝑠
𝑦
𝑣,𝐼
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑑𝑎𝑡𝑎 𝑝𝑎𝑡ℎ 𝑓𝑟𝑜𝑚 𝑣 𝑡𝑜 𝑦 𝑎𝑛𝑑 𝐼 𝑖𝑠 𝑡ℎ𝑒 𝑣𝑎𝑙𝑖𝑑𝑖𝑡𝑦 𝑡𝑖𝑚𝑒𝑠𝑝𝑎𝑛 𝑜𝑓 𝑣,
𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡
𝑥
is the
𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡, 0, 𝑛𝑜𝑤
pair where 𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡 is the root of the graph in which 𝑑𝑎𝑡𝑎 𝑛𝑜𝑑𝑒 exists and 𝑥 is a
𝑑𝑎𝑡𝑎 𝑛𝑜𝑑𝑒, 𝑖𝑛𝑡𝑒𝑟𝑣𝑎𝑙
pair, 𝑟
𝑑𝑎𝑡𝑎𝑅𝑜𝑜𝑡, 0, 𝑛𝑜𝑤
,
𝑐ℎ𝑖𝑙𝑑
𝑥
𝑣,𝐼
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑑𝑎𝑡𝑎 𝑝𝑎𝑡ℎ 𝑜𝑓 𝑙𝑒𝑛𝑔𝑡ℎ 1 𝑓𝑟𝑜𝑚 𝑥 𝑡𝑜 𝑣 𝑎𝑛𝑑 𝐼 𝑖𝑠 𝑡ℎ𝑒 𝑣𝑎𝑙𝑖𝑑𝑖𝑡𝑦 𝑡𝑖𝑚𝑒𝑠𝑝𝑎𝑛 𝑜𝑓 𝑣
𝑠𝑢𝑏𝑛𝑜𝑑𝑒𝑠
𝑦
𝑣
,𝑖,𝑣
,𝑣
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑐ℎ𝑎𝑛𝑔𝑒 𝑝𝑎𝑡ℎ 𝑓𝑟𝑜𝑚 𝑦 𝑡𝑜 𝑣
𝑎𝑛𝑑 𝑖 𝑖𝑠 𝑡ℎ𝑒 𝑡𝑖𝑚𝑒𝑠𝑡𝑎𝑚𝑝 𝑜𝑓 𝑣
𝑝𝑟𝑒𝑛𝑜𝑑𝑒𝑠
𝑦
𝑣
,𝑖,𝑣
,𝑣
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑐ℎ𝑎𝑛𝑔𝑒 𝑝𝑎𝑡ℎ 𝑓𝑟𝑜𝑚 𝑣
𝑡𝑜 𝑦 𝑎𝑛𝑑 𝑖 𝑖𝑠 𝑡ℎ𝑒 𝑣𝑎𝑙𝑖𝑑𝑖𝑡𝑦 𝑡𝑖𝑚𝑒𝑠𝑝𝑎𝑛 𝑜𝑓 𝑣
,
𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡
𝑥
is the
𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡,0,∅,∅
tuple where 𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡 is the root of the graph in which 𝑐ℎ𝑎𝑛𝑔𝑒 𝑛𝑜𝑑𝑒 exists and 𝑥
is a
𝑐ℎ𝑎𝑛𝑔𝑒 𝑛𝑜𝑑𝑒, 𝑖𝑛𝑠𝑡𝑎𝑛𝑐𝑒, 𝑑𝑎𝑡𝑎 𝑛𝑜𝑑𝑒 𝑏𝑒𝑓𝑜𝑟𝑒, 𝑑𝑎𝑡𝑎 𝑛𝑜𝑑𝑒 𝑎𝑓𝑡𝑒𝑟
tuple, 𝑟
𝑐ℎ𝑎𝑛𝑔𝑒𝑅𝑜𝑜𝑡,0,∅,∅
,
𝑐ℎ𝑖𝑙𝑑
𝑥
𝑣
,𝑖,𝑣
,𝑣
|
𝑡ℎ𝑒𝑟𝑒 𝑒𝑥𝑖𝑠𝑡𝑠 𝑎 𝑐ℎ𝑎𝑛𝑔𝑒 𝑝𝑎𝑡ℎ 𝑜𝑓 𝑙𝑒𝑛𝑔𝑡ℎ 1 𝑓𝑟𝑜𝑚 𝑥 𝑡𝑜 𝑣
𝑎𝑛𝑑 𝑖 𝑖𝑠 𝑡ℎ𝑒 𝑡𝑖𝑚𝑒𝑠𝑡𝑎𝑚𝑝 𝑜𝑓 𝑣
Evo-Path: Querying Data Evolution through Complex Changes
357

expression 𝑄
⟦
𝑞
⟧
𝑥 denotes whether or not an
evolution condition 𝑞
is satisfied. For the change
part, four similar semantic functions are defined:
𝑆
,𝑄
, 𝑄
and 𝑄
. 𝑆
⟦
𝑝
⟧
𝑥 denotes the sequence of
tuples (change-node, instance, data-node-before, data-
node-after) selected by pattern 𝑝 when 𝑥 is the
context tuple. It may also denote a sequence of values.
The boolean expression 𝑄
⟦
𝑞
⟧
𝑥 denotes whether or
not the qualifier 𝑞 is satisfied when the context tuple
(change-node, instance, data-node-before, data-node-
after) is 𝑥. The boolean expression 𝑄
⟦
𝑞
⟧
𝑥 denotes
whether or not a temporal condition 𝑞
is satisfied,
while the boolean expression 𝑄
⟦
𝑞
⟧
𝑥 denotes
whether or not an evolution condition 𝑞
is satisfied.
In Table 2 the formal semantics of the most common
evo-path constructs are presented.
For the data root and change root it holds: The
validity timespan of the data root is by definition
0, now , as it is always valid in time. The
timestamp of the change root is by definition 0, the
data-node-before and data-node-after are undefined
(∅), as it does not represent an actual change.
3.4 Implementation
In order to implement evo-path, each valid evo-path
expression is translated into an equivalent XPath
expression over evoXML. Table 3 summarizes the
translation rules. Each data/change path expression
(case A) is evaluated starting from the data/change
root. Each temporal predicate (case B) is mapped to
an XPath predicate over evoXML attributes
evo:ts,
evo:te and evo:tt. Each evolution predicate (case
C) is mapped to an XPath predicate over the
evoXML attributes
evo:before or/and evo:after.
These attributes appear on change elements and
should be equal to
evo:id attribute of data elements.
Moreover, recall that evoXML encodes evo-graph in
a top-down non-replicated approach (Stavrakas and
Papastefanatos, 2011): if a child node is pointed to by
multiple parent versions, the element corresponding
to the child node is contained in the oldest parent
element, while subsequent parent versions contain
"clone" elements of the child. These are empty
elements pointing to the "original" child element via
evo:ref attribute. This feature is handled while
translating a data path expression to an equivalent
XPath expression (case D). The returned nodes of a
data path expression should be the "original" ones,
i.e. those with an
evo:id attribute (rule 1). Similar
holds for predicates that are used to find a specific
node, e.g. based on position (rule 2). For predicates
that are used to find a node that contains a specific
value, the returned nodes should be the "original"
ones and the contained value should be checked in an
"original" child node. However, the node in context
may have either an "original" or a "clone" child node.
In the latter case, the "clone" child node is used to
access the pointed "original" one. Thus, in rule 3 two
cases are identified:
p1 is an "original" node and
contains the "original" node
p2 with value, or p1 is
an "original" node and contains the "clone" node
p2
pointing to an "original" node with value. This is
extended in rule 4 with an additional location step.
For
p3 a third case is identified: p1 is an "original"
node which contains the "original" node
p2 with
value and the "clone" node
p3, which is used to
access the "original" pointed node
p3. The case of
having
p1 as "original" node and p2 and p3 as
"clone" nodes is not identified, since it eventually
ends up to one of the rest cases. Finally, note that
XPath predicates on other node types, like attributes,
are not considered, since in evoXML evolving data
are represented on element nodes.
Below, we show the XPath expressions for the
Section 3.2 evo-path queries, generated following
the translation rules. For simplicity evo namespace
is omitted. evoXML.xml contains the evoXML
representation of evo-graph in Figure 1.
(Q1) let $d:=doc("evoXML.xml")/evo:evoXML/
evo:DataRoot
return $d//Diabetes/categories
[@evo:te!='now']
(Q2) let $d:=doc("evoXML.xml")/evo:evoXML/
evo:DataRoot,
$c:=doc("evoXML.xml")/evo:evoXML/
evo:ChangeRoot
return $c//*[@evo:before=
$d//Diabetes//*/@evo:id or
@evo:after=
$d//Diabetes//*/@evo:id]
[ .//evo:create[@evo:before=
$
d//Diabetes/categories/cat/@evo:id or
@evo:after=
$d//Diabetes/categories/cat/@evo:id] ]
(Q3) let $c:=doc("evoXML.xml")/evo:evoXML/
evo:ChangeRoot
return $c//reorg_diab_cat/*
(Q4) let $d:=doc("evoXML.xml")/evo:evoXML/
evo:DataRoot,
$c:=doc("evoXML.xml")/evo:evoXML/
evo:ChangeRoot
return $d//*[@evo:before=
$c//reorg_diab_cat//*/@evo:id]
4 RELATED WORK
An early work (Chawathe, Abiteboul and Widom
1999) proposes DOEM, an extension of OEM,
representing changes as annotations on the nodes
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
358
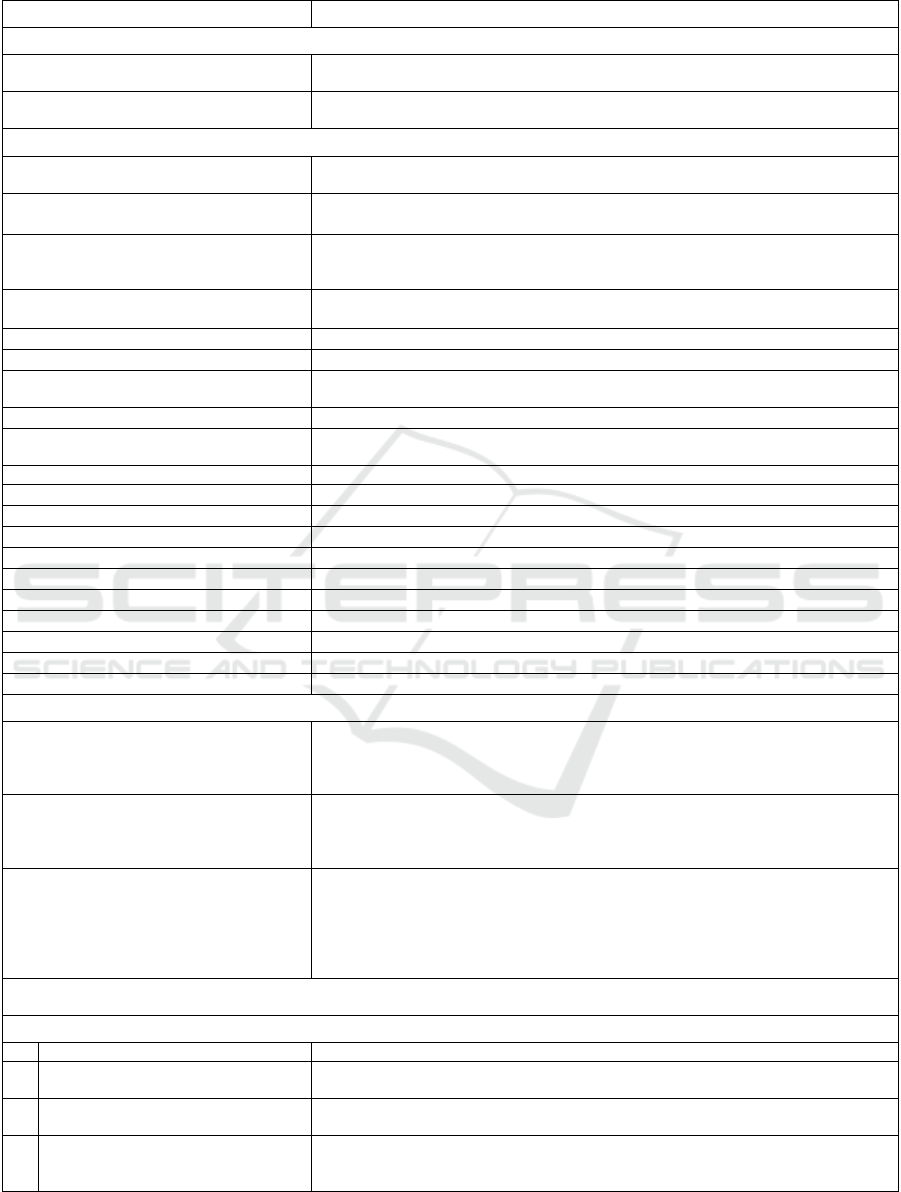
Table 3: Evo-Path to XPath translation.
Evo-Path XPath
A. Data and Change Path Expressions
data_path_expr doc("evoXML.xml")/evo:evoXML/evo:DataRoot/mapped_data_path_e
xpr
<change_path_expr> doc("evoXML.xml")/evo:evoXML/evo:ChangeRoot/mapped_change_pa
th_expr
B. Temporal Predicates
[ts() in (t_1, t_2)],where 𝑡
_
2∈ℕ
[@evo:ts>= t_1 and
(
if @evo:te='now' then false() else
@evo:te<= t_2)]
[ts() contains (t_1, t_2)],
where 𝑡
_
2∈ℕ
[@evo:ts<= t_1 and
(if @evo:te='now' then true() else @evo:te>= t_2)]
[ts() meets (t_1, t_2)],
where 𝑡
_
2∈ℕ
[if @evo:te='now' then (@evo:ts>= t_1 and @evo:ts<= t_2)
else((@evo:ts>= t_1 and @evo:ts<= t_2) or
(@evo:te>= t_1 and @evo:te<= t_2))]
[ts() equals (t_1, t_2)], where
𝑡
_
2∈ℕ
[@evo:ts = t
_
1 and (if @evo:te='now' then false() else
@evo:te = t_2)]
[ts() in (t_1, 'now')]
[@evo:ts>= t_1]
[ts() contains (t_1, 'now')]
[@evo:ts<=t_1 and @evo:te='now']
[ts() meets (t_1, 'now')]
[if @evo:te='now' then true()else (@evo:ts>=t
_
1 or
@evo:te>=t_1)]
[ts() equals (t_1, 'now')]
[@evo:ts = t_1 and @evo:te='now']
[ts() covers t], where 𝑡∈ℕ
[@evo:ts<= t and (if @evo:te='now' then true() else
@evo:te>= t)]
[ts() covers 'now'] [@evo:te='now']
[tstart() operator t], where 𝑡∈ℕ
[@evo:ts operator t]
[tend() > t], where 𝑡∈ℕ
[if @evo:te='now' then true() else @evo:te> t]
[tend() >= t], where 𝑡∈ℕ
[if @evo:te='now' then true() else @evo:te>= t]
[tend() = t], where 𝑡∈ℕ
[if @evo:te='now' then false() else @evo:te = t]
[tend() < t], where 𝑡∈ℕ
[if @evo:te='now' then false() else @evo:te< t]
[tend() <= t], where 𝑡∈ℕ
[if @evo:te='now' then false() else @evo:te<= t]
[tend() = 'now']
[@evo:te='now']
[tend()< 'now']
[@evo:te!='now']
[tend()<= 'now']
[true()]
[tt() operator t], where 𝑡∈ℕ
[@evo:tt operator t]
C. Evolution Predicates
data_path_expr
[evo-
before(<change_path_expr>)]
doc("evoXML.xml")/evo:evoXML/evo:DataRoot/data_path_expr[@ev
o:id=
doc("evoXML
.
xml")/evo:evoXML/evo:ChangeRoot/change
_
path
_
expr
/
@evo:before]
data_path_expr
[evo-
after(<change_path_expr>)]
doc("evoXML.xml")/evo:evoXML/evo:DataRoot/data_path_expr[@ev
o:id=
doc("evoXML
.
xml")/evo:evoXML/evo:ChangeRoot/change
_
path
_
expr
/
@evo:after]
data_path_expr
[evo-both(<change_path_expr>)]
doc("evoXML.xml")/evo:evoXML/evo:DataRoot/data_path_expr[@ev
o:id=
doc("evoXML
.
xml")/evo:evoXML/evo:ChangeRoot/change
_
path
_
expr
/
@evo:before or @evo:id=
doc("evoXML
.
xml")/evo:evoXML/evo:ChangeRoot/change
_
path
_
expr
/
@evo:after]
<change_path_expr [evo-filter(data_path_expr)]> where evo-filter is evo-before or evo-
a
fter
or evo-both are defined symmetrically
D. Plain Data Path Expressions
1 /p /p[@evo:id]
2 /p[position predicate] /p[(@evo:id and position predicate) or
(@evo:id=/p[position predicate]/@evo:ref)]
3 /p1[p2 op value] /p1[@evo:id and p2 op value] |
/p1[@evo:id and p2/@evo:ref=/p1[p2 op value]/p2/@evo:id]
4 /p1[p2 op value]/p3 (/p1[@evo:id and p2 op value] |
/p1[@evo:id and p2/@evo:ref=/p1[p2 op value]/p2/@evo:id] |
/p1[p3/@evo:id=/p1[p2 op value]/p3/@evo:ref])/p3[@evo:id]
Evo-Path: Querying Data Evolution through Complex Changes
359

and edges of the OEM graph. In (Marian et al.,
2001), a diff algorithm is employed for detecting
changes between two versions of an XML
document and storing them as edit scripts or deltas.
A similar approach is in (Chien, Tsotras and
Zaniolo, 2001), where a referenced-based
identification of objects is used across versions. In
(Gergatsoulis and Stavrakas, 2003) MXML, an
XML extension that uses context information to
express time and model multifaceted documents, is
proposed. Other works deal with change modelling
(Rizzolo et al., 2009) and detection (Papavassiliou
et al., 2009), (Galani, Papastefanatos and
Stavrakas, 2016) on semantic data and RDF.
In (Rizzolo and Vaisman, 2008), an XML
document is modelled as a directed graph and
transaction time is attached at the edges. In (Gao
and Snodgrass, 2003), a temporal query language
for adding valid time support in XQuery is
presented. In (Wang and Zaniolo, 2003) a
temporally grouped data model is employed for
uniformly representing and querying versions. In
(Moon et al., 2008), this technique is extended for
publishing the history of a relational database in
XML and a set of schema modification operators
(SMOs) is used for representing mappings between
successive schema versions. (Amagasa, Yoshikawa
and Uemura, 2000) deal with archiving curated
databases for scientific data, using timestamps and
merging all versions into one hierarchy. (Buneman,
Chapman and Cheney, 2006) deal with provenance
in curated databases. User actions are recorded in
sequence and stored as provenance links.
Our model introduces a change-based view for
evolving data. Changes are not derived by data
versions, but are modelled as first class citizens
along with data. Changes are not described via
diffs or transformations with edit scripts between
versions, but are complex objects operating on
data, exhibiting structural, semantic, and temporal
properties. Thus, querying evolution involves
searching on both data and change structure, using
temporal- and change-based conditions. Change-
centric modelling can provide additional
information on what, why, and how data evolved.
5 CONCLUSIONS
In this paper, we formally defined evo-path: a
language for querying evolving data and changes
in a uniform way. Evo-path operates on evo-graph,
a model that captures data versions and structured
changes. We also defined evo-path translation into
plain XPath expressions, which are evaluated on
evoXML, an XML representation of evo-graph.
Our next steps involve experimenting evo-path.
ACKNOWLEDGEMENTS
This research has been funded by the project
"Moving from Big Data Management to Data
Science" (MIS 5002437/3)-Action "Reinforcement
of the Research and Innovation Infrastructure"
(funded by Greece and the European Regional
Development Fund).
REFERENCES
Amagasa, T., Yoshikawa, M., Uemura, S. (2000). A
Data Model for Temporal XML Documents. In
DEXA.
Buneman, P., Khanna, S., Tajima, K., Tan, W.C. (2004).
Archiving Scientific Data. In ACM Transactions on
Database Systems, Vol. 20, pp 1-39.
Buneman, P., Chapman, A. P., Cheney, J. (2006).
Provenance Management in Curated Databases. In
SIGMOD'06.
Chawathe, S., Abiteboul, S., Widom, J. (1999).
Managing Historical Semistructured Data. In
Journal of Theory and Practice of Object Systems,
Vol. 24(4), pp.1-20.
Chien, S-Y., Tsotras, V. J., Zaniolo, C. (2001). Efficient
Management of Multiversion Documents by Object
Referencing. In VLDB.
Gao, D., Snodgrass, R. T. (2003). Temporal Slicing in
the Evaluation of XML Queries. In VLDB.
Gergatsoulis, M., Stavrakas, Y. (2003). Representing
Changes in XML Documents using Dimensions. In
1st International XML Database Symposium.
Marian, A., Abiteboul, S., Cobena, G., Mignet, L.
(2001). Change-Centric Management of Versions in
an XML Warehouse. In VLDB.
Moon, H.J., Curino, C., Deutsch, A., Hou, C.Y.,
Zaniolo, C. (2008). Managing and querying
transaction-time databases under schema evolution.
In VLDB.
National research council - Committee on Frontiers at
the Interface of Computing and Biology (2005).
Catalyzing Inquiry at the Interface of Computing and
Biology. Edited by J. C. Wooley, H. S. Lin.,
National Academies Press.
Papavassiliou, V., Flouris, G., Fundulaki, I., Kotzinos,
D., Christophides, V. (2009). On Detecting High-
Level Changes in RDF/S KBs. In ISWC.
Rizzolo, F., Vaisman, A. A. (2008). Temporal XML:
modeling, indexing, and query processing. In VLDB
J., 17(5): 1179-1212.
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
360

Rizzolo, F., Velegrakis, Y., Mylopoulos, J., Bykau, S.
(2009). Modeling Concept Evolution: a Historical
Perspective. In ER.
Stavrakas, Y., Papastefanatos, G. (2010). Supporting
Complex Changes in Evolving Interrelated Web
Databanks. In CoopIS.
Stavrakas, Y., Papastefanatos, G. (2011). Using
Structured Changes for Elucidating Data Evolution.
In DaLi (with ICDE 2011).
Papastefanatos, G., Stavrakas, Y., Galani, T. (2013).
Capturing the History and Change Structure of
Evolving Data. In DBKDA.
Galani, T., Papastefanatos, G., Stavrakas, Y. (2016). A
language for defining and detecting interrelated
complex changes on RDF(S) knowledge bases. In
ICEIS.
Wadler, P. (1999). A Formal Semantics of Patterns in
XSLT. In Markup Technologies.
Wang, F., Zaniolo, C. (2003). Temporal Queries in XML
Document Archives and Web Warehouses. In TIME.
Robie, J., Dyck, M., Spiegel, J. (2017, March 21). XML
Path Language (XPath) 3.1 W3C Recommendation.
https://www.w3.org/TR/2017/REC-xpath-31-
20170321/.
Evo-Path: Querying Data Evolution through Complex Changes
361
