
Making Data Big for a Deep-learning Analysis: Aggregation of Public
COVID-19 Datasets of Lung Computed Tomography Scans
Francesca Lizzi
1,2
, Francesca Brero
3,4
, Raffaella Fiamma Cabini
3,5
, Maria Evelina Fantacci
2,6
,
Stefano Piffer
7,8
, Ian Postuma
3
, Lisa Rinaldi
3,4
and Alessandra Retico
2
1
Scuola Normale Superiore, Pisa, Italy
2
National Institute for Nuclear Physics (INFN), Pisa Division, Pisa, Italy
3
INFN, Pavia Division, Pavia, Italy
4
Department of Physics, University of Pavia, Pavia, Italy
5
Department of Mathematics, University of Pavia, Pavia, Italy
6
Department of Physics, University of Pisa, Pisa, Italy
7
Department of Biomedical Experimental Clinical Science ”M. Serio”, University of Florence, Florence, Italy
8
INFN, Florence Division, Florence, Italy
Keywords:
COVID-19, Lung CT, U-net, Data Aggregation, Image Segmentation.
Abstract:
Lung Computed Tomography (CT) is an imaging technique useful to assess the severity of COVID-19 in-
fection in symptomatic patients and to monitor its evolution over time. Lung CT can be analysed with the
support of deep learning methods for both aforementioned tasks. We have developed a U-net based algorithm
to segment the COVID-19 lesions. Unfortunately, public datasets populated with a huge amount of labelled
CT scans of patients affected by COVID-19 are not available. In this work, we first review all the currently
available public datasets of COVID-19 CT scans, presenting an extensive description of their characteristics.
Then, we describe the design of the U-net we developed for the automated identification of COVID-19 lung
lesions. Finally, we discuss the results obtained by using the different publicly available datasets. In particu-
lar, we trained the U-net on the dataset made available within the COVID-19 Lung CT Lesion Segmentation
Challenge 2020, and we tested it on data from the MosMed and the COVID-19-CT-Seg datasets to explore the
transferability of the model and to assess whether the image annotation process affects the detection perfor-
mances. We evaluated the performance of the system in lesion segmentation in terms of the Dice index, which
measures the overlap between the ground truth and the predicted masks. The proposed U-net segmentation
model reaches a Dice index equal to 0.67, 0.42 and 0.58 on the independent validation sets of the COVID-
19 Lung CT Lesion Segmentation Challenge 2020, on the MosMed and on the COVID-19-CT-Seg datasets,
respectively. This work focusing on lesion segmentation constitutes a preliminary work for a more accurate
analysis of COVID-19 lesions, based for example on the extraction and analysis of radiomic features.
1 INTRODUCTION
Lung Computed Tomography (CT) is a very sensitive
medical imaging technique to detect lung lesions due
to COVID-19. It can be used for the diagnosis, prog-
nosis and for monitoring the disease evolution over
time. Despite the use of CT for diagnosis is not rec-
ommended by the World Health Organization (World
Health Organization, 2020), lung CT analysis can be
very informative regarding the severity of the disease
and its time evolution (Fang et al., 2021). The use
of CT in clinical practice for COVID-19 diagnosis
in symptomatic patients has been explored. Since
the unexpected outbreak of the pandemic, physicians
tried to use CT imaging of the chest to diagnose
COVID-19 disease. The first publication describing
in details radiological findings of CT was published
in January, the 24 of 2020 (Huang et al., 2020) and it
describes the radiological findings of the majority of
COVID-19 hospitalized patients of this study, such as
bilateral multiple lobular and subsegmental areas of
consolidation and bilateral ground-glass opacity. Af-
terwards, several studies have been published to de-
scribe the radiological findings of COVID-19 chest
CT (Carotti et al., 2020). A summary of all possible
findings and their incidence is reported in Table 1.
316
Lizzi, F., Brero, F., Cabini, R., Fantacci, M., Piffer, S., Postuma, I., Rinaldi, L. and Retico, A.
Making Data Big for a Deep-learning Analysis: Aggregation of Public COVID-19 Datasets of Lung Computed Tomography Scans.
DOI: 10.5220/0010584403160321
In Proceedings of the 10th International Conference on Data Science, Technology and Applications (DATA 2021), pages 316-321
ISBN: 978-989-758-521-0
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
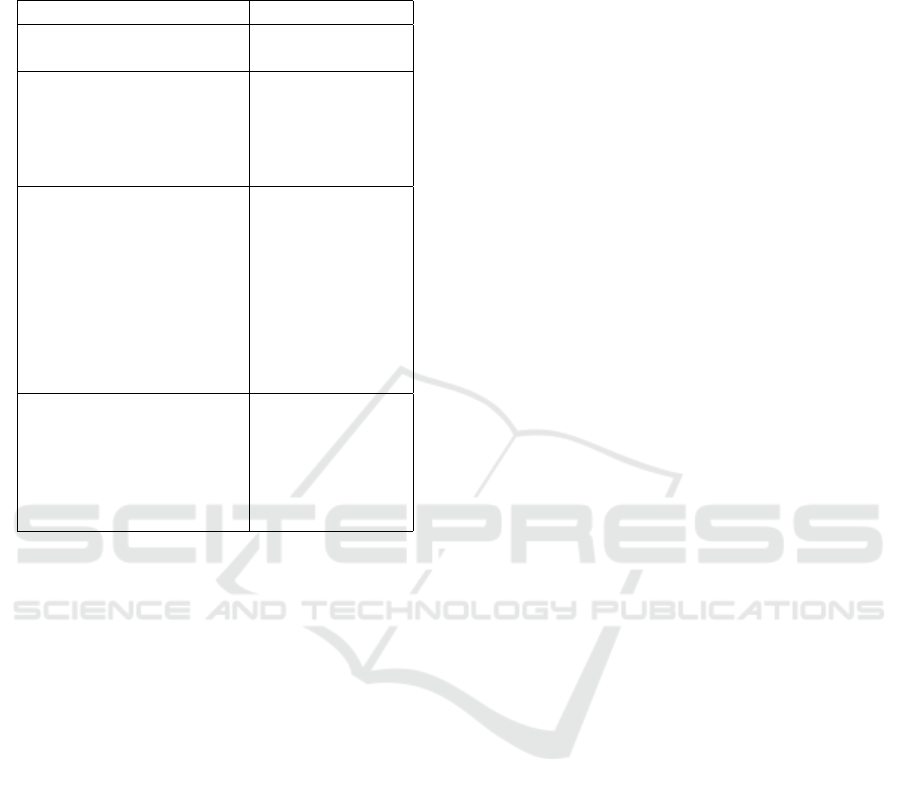
Table 1: Summary of COVID-19 chest CT findings and
their incidence on the population. The normal chest CT
findings are also associated to symptomaticity (Huang et al.,
2020).
Findings Incidence
Normal chest CT findings 10.6% (95%
CI: 7.6%, 13.7%)
Ground-Glass opacity,
Lower lobe involvement, High incidence
Bilateral abnormalities, (More than 70%)
Vascular enlargement,
Posterior predilection,
Consolidation, linear
opacity, septal thickening
and/or reticulation,
crazy-paving pattern, Intermediate
air bronchogram, pleural incidence
thickening, halo sign, (between 70%
bronchiectasis, nodules, and 10%)
bronchial wall thickening,
reversed halo sign
Pleural effusion,
lymphadenopathy,
tree-in-bud sign, Low incidence
central lesion distribution, (less than 10%)
pericardial effusion,
cavitating lung lesions
We underline that the dataset used by (Huang
et al., 2020) contains a very limited number of CT
scans (41 patients) and it is private. Most of the chest
CT findings cannot be related exclusively to COVID-
19 because they are nonspecific signs of disease and
they are strongly related to the stage of the disease.
This means that there are other forms of pneumonia
that may have the same signs such as SARS-CoV-1
and MERS-CoV. For this reason, the World Health
Organization (WHO) defined as “confirmed case” the
patient that have been tested positive for COVID-
19 RT-PCR, irrespective of clinical signs and symp-
toms (World Health Organization, 2020). Further-
more, it is necessary to differentiate the COVID-19
infections not only from other viral pneumonia but
also from bacterial pneumonia, such as mycoplasma
pneumonia (Ishiguro et al., 2019). The use of chest
CT to diagnose COVID-19 is, hence, under discus-
sion since it implies the use of ionizing radiation (Sci-
entiae et al., 2020) while its ability to monitor the pro-
gression of the disease seems to be a promising way
to use lung CTs (Adams et al., 2020). Artificial In-
telligence (AI) is a powerful instrument that allows to
analyse a huge quantity of data, such as CT scans and,
hence, it can be used to monitor and study COVID-
19 CT signs (G
¨
ulbay et al., 2021). Unfortunately,
AI implementations require a great amount of data,
which may be not easily available. This is especially
true when deep-learning methods are used. Since
the beginning of the pandemic, some lung CT scans
of COVID-19 patients have been released by differ-
ent institutions following different guidelines for both
image acquisition and annotation (ground truth). In
this work, all the public lung COVID-19 CT datasets,
to the best of our knowledge, suitable for training
AI-based systems are reviewed. In this work, we
present an extensive description of the currently pub-
licly available datasets, which present different char-
acteristics, and discuss the segmentation results ob-
tained by using them. In particular, we trained a U-net
on the dataset released within the COVID-19 Lung
CT Lesion Segmentation Challenge 2020 (An et al.,
2020) and tested it on data from MosMed (Moro-
zov et al., 2020) and COVID-19-CT-Seg datasets (Ma
et al., 2020). Finally, the limits and the advantages of
aggregating this kind of data are discussed.
2 AI AND MEDICAL IMAGE
DATASET ISSUES
AI has been used to analyse and process CT to diag-
nose COVID-19, to segment lesions inside the lungs
and, also, in longitudinal studies to track the evolution
of the disease (Ma et al., 2020). AI based methods,
especially deep-learning ones, need a huge amount of
labelled data that are not easy to collect and share.
As already described in the introduction, some stud-
ies use private datasets which do not allow a fair com-
parison with other AI based systems. Furthermore,
the characteristics of CT images depend on the scan-
ner, on the acquisition and the reconstruction proto-
cols and on other information which may not be avail-
able. This can be due also to the anonymization pro-
cess needed to preserve subjects’ privacy or to the
use of image format different from DICOM (Stan-
dard DICOM, 2021), such as the NIfTI format. DI-
COM is the most used image format for medical im-
ages and it contains several meta-data in its header.
The DICOM header stores many information, some
of which is Protected Health Information (PHI) or pri-
vate keys that are inserted and encoded by the manu-
facturer and may contain PHI as well. On the other
hand, some meta-data, such as anode characteristics
or X-ray parameters, do not contain PHI and they can
be useful in analysing images. For all these reasons,
anonymizing a DICOM file is not a trivial problem
and dataset may include images in a different format
such as NIfTI (Moore et al., 2015). Deep learning
based methods often require the association with a la-
Making Data Big for a Deep-learning Analysis: Aggregation of Public COVID-19 Datasets of Lung Computed Tomography Scans
317

bel depending on the task we want to solve. Many ap-
proaches are based on supervised learning and, hence,
image annotation plays a crucial role. Usually, medi-
cal image labels are given by one or more radiologists
with experience in the specific field, and image an-
notation is a very time-consuming task. This is the
reason why there is a general lack of public labelled
datasets of medical images. In order to save time, it
may happen that the labelling is made with the sup-
port of an automatic tool and then labels are adjusted
manually by one or more physicians.
3 LUNG CT DATASETS
In this section, the currently available datasets of
lung CT and their annotation process are reported.
The dataset are: COVID-19 Lung CT Lesion Seg-
mentation Challenge 2020 Dataset, MosMed Dataset,
COVID-19-CT-Seg Dataset and TCIA-COVID-19-
AR.
3.1 COVID-19 Lung CT Lesion
Segmentation Challenge 2020
Dataset
The COVID-19 Lung CT Lesion Segmentation Chal-
lenge 2020 (Challenge dataset) dataset is a pub-
lic dataset made by 199 unenhanced chest CT
with positive RT-PCR for SARS-CoV-2 patients (An
et al., 2020), published as training set in the occa-
sion of the COVID GrandChallenge (https://covid-
segmentation.grand-challenge.org/). Each CT is an-
notated voxel-wise and indicates all the COVID-19
lesions in a unique mask. Data has been provided
by The Multi-national NIH Consortium for CT AI in
COVID-19 via the NCI TCIA public website in NIfTI
format. Annotations have been made using a COVID-
19 segmentation model provided by NVIDIA that
takes a full CT chest volume and produces pixel wise
segmentation masks of COVID-19 lesions. These
segmentation masks have been adjusted manually by
a board of certified radiologists in order to give 3D
consistency to the lesion masks. The annotations of
the training set have been published in the context of
the challenge while the system performance has been
evaluated by challenge organizers on an independent
validation set of 50 CT scans, for which the lesion an-
notations were not publicly released. A third set, an
independent test set consisting of 46 CT scans, was
used to the define the final ranking among the partic-
ipants, and, also in this case, the lesion segmentation
annotations were not publicly released.
3.2 MosMed Dataset
MosMed (Morozov et al., 2020) is a dataset of
COVID-19 Chest CT scans collected by the Re-
search and Practical Clinical Center for Diagnos-
tics and Telemedicine Technologies of the Moscow
Health Care Department. It includes 1110 CT stud-
ies taken from 1110 patients and it is provided with a
labelling that consists of 5 classes, based on the per-
centage of involved lung parenchyma. A small sub-
set of class CT-1 cases (50 patients) has been anno-
tated by expert radiologists with the support of Med-
Seg software (2020 Artificial Intelligence AS). The
image annotations consist of binary masks in which
white voxels represent both Ground-Glass opacities
and consolidation. Both CT scans and annotations
were provided in NIfTI format. During the DICOM-
to-NIfTI conversion only one every 10th image was
preserved (MosMed, 2020).
3.3 COVID-19-CT-Seg Dataset
The COVID-19-CT-Seg dataset is a collection of CT
scans made available by the Coronacases Initiative
and Radiopaedia (Ma et al., 2020) and contains 20
CT scans of patients resulted positive for RT-PCR
COVID-19 infection. It is a public dataset which con-
tains annotations related to both lung and infection
localization. The ground truth has been made in three
steps: first, junior radiologists (1-5 years of experi-
ence) delineated the annotations of lungs and infec-
tions, then two radiologists (5-10 years of experience)
refined the labels and finally the annotations were ver-
ified and optimized by a senior radiologist (more than
10 years of experience in chest radiology). The anno-
tations have been produced with ITK-SNAP software.
Ten cases of this dataset were provided in 8-bit depth
which are not commonly used in clinical practice.
3.4 TCIA-COVID-19-AR
The TCIA-COVID-19-AR (Desai et al., 2020) is
a dataset of COVID-19 cases taken from a rural
population, which is often underrepresented in public
datasets. It contains 24 CT scans of patients with
both lung lesions due to COVID-19 and control
cases. Each patient is described by a set of clinical
data correlates that includes key radiology findings.
Moreover, for each patient the information about In-
tensive Care Unit (ICU) admission is included while
annotations on images are not included in this dataset.
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
318

Figure 1: U-net summary: the U-shaped neural network is
made of 5 levels of depth. In the left path (compression), the
input is processed through convolutions, activation layers
(ReLu) and instance normalization layers, while in the right
one (decompression), in addition to those already men-
tioned, 3D Transpose Convolution (de-convolution) layers
are also introduced. Each block (green) is made of 3 convo-
lutional layers.
4 COVID-19 LESION
SEGMENTATION
We developed an automatic system which can seg-
ment COVID-19 lesions based on a U-net (Ron-
neberger et al., 2015) in the framework of the
COVID-19 Lung CT Lesion Segmentation Challenge
2020 (GrandChallenge, 2020). First, a bounding box
which contains the lungs has been built for each CT
scan to reduce as much as possible the background
from the images. An in-house lung segmentation al-
gorithm based on active contours was developed for
this purpose and implemented in matlab (The Math-
Works, Inc.). This algorithm, which accurately seg-
ments the lung parenchyma in absence of lesions,
has very limited performance on CT scans of sub-
jects with COVID-19 lesions. The CT images have
been cropped to the bounding boxes, resized to a ma-
trix of 200x150x100 voxels and a CT windowing in
[-1000,300] range of Hounsfield Units has been ap-
plied on them to enhance the COVID-19 lesions. A
schematic representation of the used U-net is reported
in Figure 1.
We trained the network on the Challenge train-
ing dataset of 199 CT scans, using a weighted cross-
entropy as loss function, and we tested it on the Chal-
lenge validation set (independent from the training
set). In order to have a sufficient number of sam-
ples, we applied data augmentation with rotations,
zooming and elastic transformation to the training set.
We tested the network also on the 50 annotated cases
of MosMed and on the 10 annotated cases of the
COVID-19-CT-Seg-Dataset. The MosMed dataset
contains images and labels taken in a very different
way with respect to those of the Challenge dataset.
The COVID-19-CT-Seg dataset has been built in a
more similar way to the Challenge one for both data
characteristics, such as slice thickness, and labelling
process. We evaluated the segmentation performance
of the trained network model in terms of Dice index
(Equation 1) defined as:
Dice
metric
=
2 · |M
true
∩ M
predict
|
|M
true
| + |M
pred
|
(1)
where M
true
is the ground truth mask and M
pred
is the
predicted one.
We participated in the challenge, obtaining a
Dice index equal to 0.67 on the challenge validation
set (GrandChallenge, 2020). Then, we computed the
segmentation performance of the trained model on the
MosMed dataset obtaining a Dice of 0.42, and on the
COVID-19-CT-Seg-Dataset, obtaining a Dice of 0.58.
We show in Figure 2 a visual comparison between
the reference COVID-19 lesion masks and the ones
predicted by the trained U-net for a representative CT
scan of the MosMed and of the COVID-19-CT-Seg
dataset.
5 DISCUSSION AND
CONCLUSIONS
We obtained good results in terms of the Dice index
as regards the segmentation of the lung lesions re-
lated to COVID-19 infection on the Challenge dataset
compared to literature (Ma et al., 2020). The re-
sults obtained on the other two datasets are not good
as the first one. We underline that on the dataset
more similar to the Challenge one, the COVID-19-
CT-Seg dataset, we obtained better results compared
to MosMed. As expected, we conclude that aggregat-
ing data from different sources can be difficult if la-
belling has been performed using different guidelines.
In fact, medical images have many parameters to be
considered, such as the resolution of pixels and the
size of the Field Of View (FOV). These parameters
can be studied in order to attempt a standardization
of images from different datasets, by contrast, differ-
ent annotation styles can not be easily standardized.
Since CT image characteristics can be variable, deep
learning is a useful method to analyse them and their
aggregation. Moreover, U-nets allows a quantification
of the volumes of both COVID-19 lesions and lungs.
On the other hand, the use of deep learning based
methods requires a huge amount of homogeneous or
harmonized data both to carry out an optimal training
Making Data Big for a Deep-learning Analysis: Aggregation of Public COVID-19 Datasets of Lung Computed Tomography Scans
319
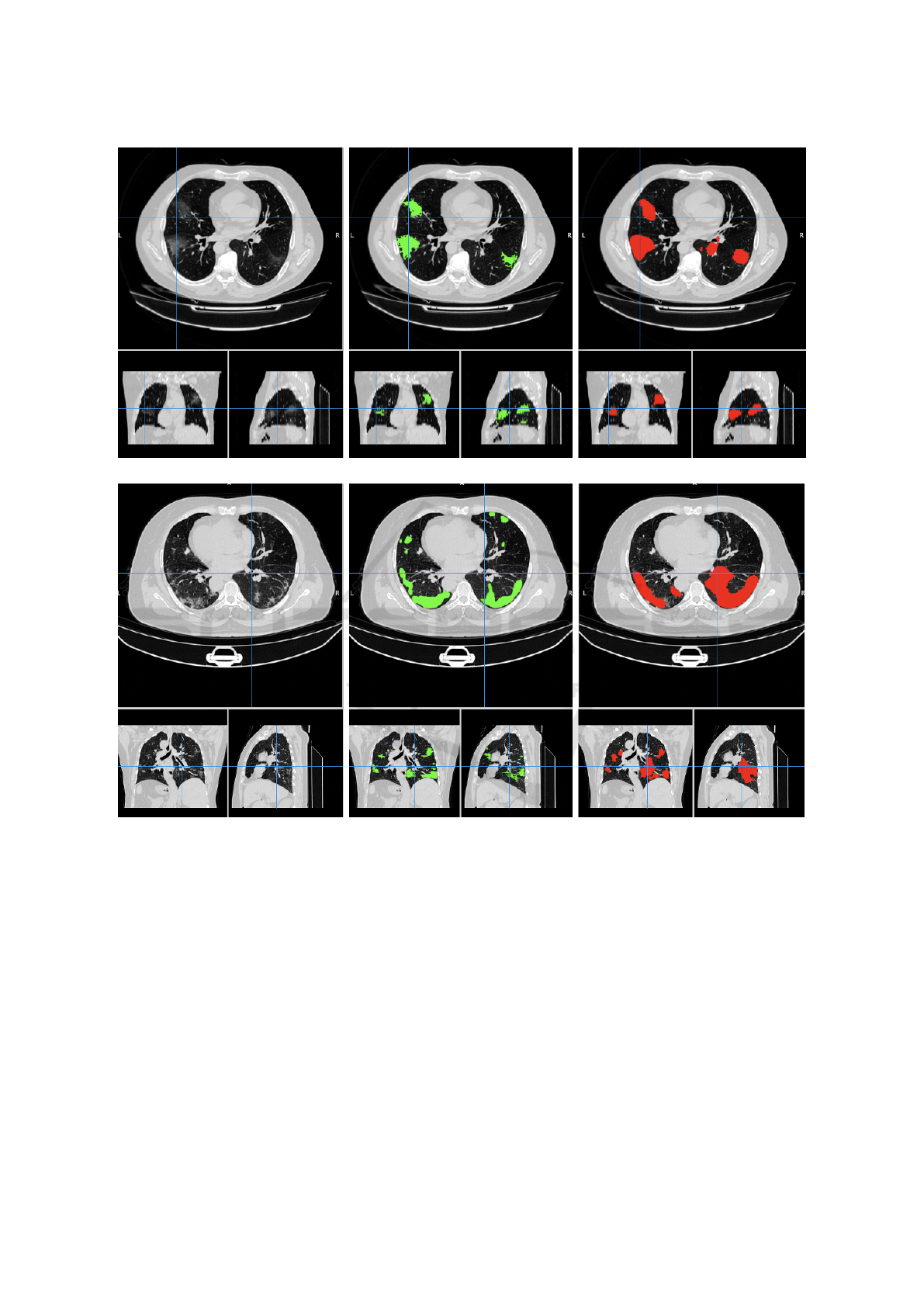
Figure 2: Visual comparison between the reference COVID-19 lesion masks (green) and the ones predicted (red) by the
trained U-net for a representative CT scan of the MosMed (first row, study-0255.nii) and of the COVID-19-CT-Seg (second
row, coronacases-001.nii) datasets. The original CT scans are shown on the left as a reference.
process and to implement a fair representation of the
population to be studied.
This preliminary study has been useful to un-
derstand which parameters should be considered as
the most critical ones in training a neural network
model. Lesion labeling and data selection criteria
are crucial for this kind of segmentation problems be-
cause of the lack of largely populated public datasets,
impacting in a relevant way on the performances.
In conclusion, we reviewed all the public available
datasets (at the best of our knowledge in April 2021),
i.e. COVID-19 Lung CT Lesion Segmentation Chal-
lenge 2020, MosMed, COVID-19-CT-Seg Dataset
and TCIA-COVID-19-AR. We used the Challenge
data to train and evaluate a U-net for COVID-19 lung
lesion segmentation, and we carried out an indepen-
dent test of the MosMed and the COVID-19-CT-Seg
datasets, obtaining good performances, as compared
to other results available in literature (Ma et al., 2020).
We are going to improve our system by adding a
module for lung segmentation which could help in
quantifying the percentage of lung tissue affected by
COVID-19 lesions. We also plan to let radiologists
evaluate the application of this algorithm on a part of
public CT datasets without labelling. Furthermore,
segmentation of COVID-19 lesions is a starting point
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
320

for an accurate radiomic analysis for the prediction,
based on radiological signs, of the clinical outcome
of patients affected by COVID-19 pneumonia.
ACKNOWLEDGEMENTS
This work has been carried out within the
Artificial Intelligence in Medicine (AIM)
project funded by INFN (CSN5, 2019-2021),
https://www.pi.infn.it/aim. We are grateful to the
staff of the Data Center of the INFN Division of
Pisa. We thank the CINECA Italian computing
center for making available part of the computing
resources used in this paper; in particular, Dr. Tom-
maso Boccali (INFN, Pisa) as PI of PRACE Project
Access #2018194658 and a 2021 ISCRA-C grant.
Moreover, we thank the EOS cluster of Department
of Mathematics ”F. Casorati” (Pavia) for computing
resources.
REFERENCES
Adams, H. J., Kwee, T. C., Yakar, D., Hope, M. D., and
Kwee, R. M. (2020). Chest CT Imaging Signature of
Coronavirus Disease 2019 Infection: In Pursuit of the
Scientific Evidence. Chest, 158(5):1885–1895.
An, P., Xu, S., Harmon, S. A., Turkbey, E. B., Sanford,
T. H., Amalou, A., Kassin, M., Varble, N., Blain,
M., Anderson, V., Patella, F., Carrafiello, G., Turkbey,
B. T., and Wood, B. J. (2020). CT Images in COVID-
19.
Carotti, M., Salaffi, F., Sarzi-Puttini, P., Agostini, A.,
Borgheresi, A., Minorati, D., Galli, M., Marotto,
D., and Giovagnoni, A. (2020). Chest CT features
of coronavirus disease 2019 (COVID-19) pneumo-
nia: key points for radiologists. Radiologia Medica,
125(7):636–646.
Desai, S., Baghal, A., Wongsurawat, T., Al-Shukri, S.,
Gates, K., Farmer, P., Rutherford, M., Blake, G.,
Nolan, T., Powell, T., Sexton, K., Bennett, W., and
Prior, F. (2020). Data from Chest Imaging with Clin-
ical and Genomic Correlates Representing a Rural
COVID-19 Positive Population [Data set].
Fang, X., Kruger, U., Homayounieh, F., Chao, H., Zhang, J.,
Digumarthy, S. R., Arru, C. D., Kalra, M. K., and Yan,
P. (2021). Association of AI quantified COVID-19
chest CT and patient outcome. International Journal
of Computer Assisted Radiology and Surgery.
GrandChallenge (2020). COVID-19 Lung CT Le-
sion Segmentation Challenge - 2020, https://covid-
segmentation.grand-challenge.org/COVID-19-20/.
G
¨
ulbay, M.,
¨
Ozbay, B. O., Mendi, B. A. R., Bas¸tu
˘
g,
A., and Bodur, H. (2021). A CT radiomics analy-
sis of COVID-19-related ground-glass opacities and
consolidation: Is it valuable in a differential diag-
nosis with other atypical pneumonias? PloS one,
16(3):e0246582.
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y.,
Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu,
T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H.,
Liu, M., Xiao, Y., Gao, H., Guo, L., Xie, J., Wang,
G., Jiang, R., Gao, Z., Jin, Q., Wang, J., and Cao,
B. (2020). Clinical features of patients infected with
2019 novel coronavirus in Wuhan, China. The Lancet,
395(10223):497–506.
Ishiguro, T., Kobayashi, Y., Uozumi, R., Takata, N.,
Takaku, Y., Kagiyama, N., Kanauchi, T., Shimizu,
Y., and Takayanagi, N. (2019). Viral pneumonia
requiring differentiation from acute and progressive
diffuse interstitial lung diseases. Internal Medicine,
58(24):3509–3519.
Ma, J., Wang, Y., An, X., Ge, C., Yu, Z., Chen, J., Zhu,
Q., Dong, G., He, J., He, Z., Nie, Z., and Yang, X.
(2020). Towards Efficient COVID-19 CT Annotation:
A Benchmark for Lung and Infection Segmentation.
pages 1–7.
Moore, S. M., Maffitt, D. R., Smith, K. E., Kirby, J. S.,
Clark, K. W., Freymann, J. B., Vendt, B. A., Tarbox,
L. R., and Prior, F. W. (2015). De-identification of
medical images with retention of scientific research
value. Radiographics, 35(3):727–735.
Morozov, S. P., Andreychenko, A. E., Pavlov, N. A.,
Vladzymyrskyy, A. V., Ledikhova, N. V., Gom-
bolevskiy, V. A., Blokhin, I. A., Gelezhe, P. B., Gon-
char, A. V., and Chernina, V. (2020). MosMedData:
Chest CT Scans with COVID-19 Related Findings
Dataset. medRxiv, page 2020.05.20.20100362.
MosMed (2020). MosMed dataset website,
https://mosmed.ai/en/.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical im-
age segmentation. Lecture Notes in Computer Sci-
ence (including subseries Lecture Notes in Artificial
Intelligence and Lecture Notes in Bioinformatics),
9351:234–241.
Scientiae, D. C.-S., Adams, H. J. A., Kwee, T. C., Hope,
M. D., Kwee, R. M., Hja, A., Tc, K., and Yakar, D.
(2020). Systematic Review and Meta- in the Diagno-
sis of Coronavirus. (December):1342–1350.
Standard DICOM (2021). DICOM standard.
World Health Organization (2020). WHO Interim guidance
20 March 2020 - Global Surveillance for COVID-19
disease caused by human infection with novel coron-
avirus (COVID-19). Who, (January):1–4.
Making Data Big for a Deep-learning Analysis: Aggregation of Public COVID-19 Datasets of Lung Computed Tomography Scans
321
