
Towards Automatic Detection and Quantification of Mildew on
Grape Leaf Disks
Razib Iqbal, Kyle Sargent and Laszlo Kovacs
College of Natural and Applied Sciences, Missouri State University, Springfield, MO, U.S.A.
Keywords: Background Removal, Downy Mildew, Grape Leaf, HSV Masking, Image Analysis.
Abstract: Downy and powdery mildews are the most serious diseases of the grapevine. A sustainable way to control
these pathogens is the breeding and deployment of resistant grape cultivars. For breeding efforts to be
effective, accurate quantification of the resistance phenotype is essential. In this paper, we present a computer-
based image recognition, processing, and analysis technique for enhancing the detection and quantification of
Plasmopara viticola and Erysiphe necator the causal agents of downy and powdery mildew, respectively. We
propose a multi-step approach that utilizes background removal and Hue-Saturation-Value (HSV) masking
as opposed to multi-faceted color channel breakdowns, photo texture evaluations, or classification-based
algorithms for the detection of mildew. Our experimental results show that our method provides reliable
results and fast performance.
1 INTRODUCTION
Plants can be classified based on two distinctions of
infection, namely, non-infected (or normal) and
infected (Awate et al, 2015). In the infected plants
category, the growth of pathogen on plants is a major
problem in the agricultural industry. To prevent it,
many cultivators turn to harmful pesticides to
slow/prevent the infection of it. While this practice is
effective, it has many drawbacks. Instead, biologists
have looked into breeding the plants selectively in
order to breed samples that are naturally resistant to
certain pathogens. In order to determine success in
this manner, we need to analyze infected samples and
determine the rate and amount of growth of infection
on those samples. In this paper, we focus on grape leaf
disks and the methods for detection and quantification
of the mildew at both the microscopic level and
human eye-level.
The existing methods for detecting mildew
include color-space analysis, texture analysis, support
vectors, and visual analysis (Awate et al, 2015;
Sandika et al, 2016; Li et al, 2011; Vijayakumar,
2012). Hardware-based image analyses, such as
(Cruz et al, 2016), rely on the capabilities of the
hardware and the cost of the hardware is a factor in
determining the aspects of the analysis. In
comparison, visual analysis even though the most
accessible and cost-efficient detection method has
factors of bias from human perception. Its primary
use is when quick and non-accurate readings are
required to give a baseline for further analysis at a
later point. Since this method is often accompanied
by result variation, we have turned to computer-based
image analysis for reliable and deterministic output
that is useful to the end user.
Color space analysis can be further divided into
multiple different categories, such as RGB color-
space analysis, BGR color-space analysis and Hue-
Saturation-Value (HSV) color space analysis. As per
(Vijayakumar, 2012), the RGB color-space can be
split between the individual color channels to point
out anomaly values caused by the growth of mildew.
This method allows for a histogram approach, which
accompanies calculating the mean value of each color
channel and tracking changes in said values. HSV and
BGR color spaces, also maintain the abilities from the
RGB color-space analysis technique. However,
creating a histogram of all colors in a single image
can be very cumbersome on a machine depending on
two factors: image quality and image resolution. Due
to this, we elected to use color space masking to
alleviate the necessity of histogram creation or any
other expensive color channel tracking approaches.
Our proposed approach tends to provide a reliable
method for quantifying the mildew growth on grape
leaves.
Iqbal, R., Sargent, K. and Kovacs, L.
Towards Automatic Detection and Quantification of Mildew on Grape Leaf Disks.
DOI: 10.5220/0010583900810086
In Proceedings of the 18th International Conference on Signal Processing and Multimedia Applications (SIGMAP 2021), pages 81-86
ISBN: 978-989-758-525-8
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
81
The rest of the paper is organized as follows: In
Section 2, we present our proposed approach with the
implementation details. In Section 3, we present our
performance evaluation. Finally, in Section 4, we
conclude this paper with our observation followed by
our future plans.
2 PROPOSED APPROACH
In order to measure the rate of mildew on the disk leaf
samples we apply Background Removal (BR) and
HSV masking to eliminate non-mildew spots from
each photo samples. We then use the Laplacian of
Gaussian (LoG) blob detection algorithm to quantify
the amount of mildew contained in each photo. We
explain these steps in the following subsections.
2.1 Image Acquisition
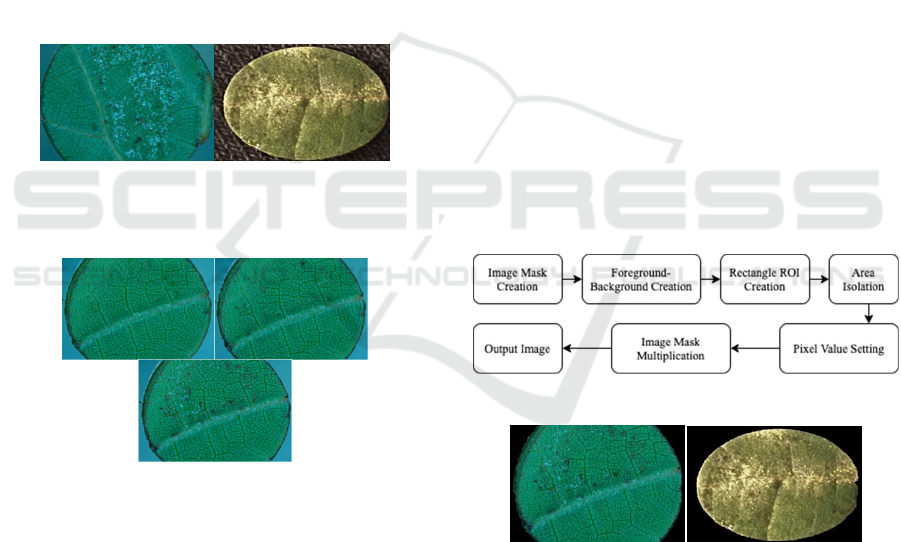
Figure 1: Photos of leaf disks infected with downy-and
powdery-mildew at the micro- (left) and macroscopic-level
(right).
Figure 2: Single downy mildew-inoculated leaf disk over
six-day span (left, right, bottom: 2dpi, 4dpi, 6dpi).
For our research, grape plants are cultivated as normal
and given time to grow into mature vines. Once the
plant has reached this stage, circular disks are cut
from the plant’s leaves, thus, supplying a grape leaf
disk. These disks are then inoculated with P. viticola
or E. necator as shown in Figure 1, and tracked over
a six-day period. Throughout these six days, the
growth of the mildew is tracked and photographs of
the leaf disks at the macroscopic level are taken in the
lab setting. On the sixth day, the resistance to the
mildew is determined based on the extent of growth,
which inversely related to disease resistance. During
this process, visual analysis is conducted to deduce a
baseline approximation of mildew on the leaf disk
and resistance to the mildew.
The downy mildew images we used for this research
were acquired from the laboratory of Dr. Lance
Cadle-Davidson at the USDA Agricultural Research
Service (Bierman et al, 2019). Our database included
53 photos in total with 48 of them being at the
microscopic level and 5 being at the human eye level.
The 43 microscopic photos span from 2-, 4-, and 6-
days past inoculation (dpi) of the leaf disk with the
mildew and photos were taken under lab-grade
microscopes and photo equipment with generally
consistent lighting placement of leaf disks within the
photos as shown in Figure 2. The macroscopic photos
of powdery mildew infected leaf disks were taken
with a USB camera with varying days past
inoculation and varying distances from the disk under
consistent lighting condition.
2.2 Background Removal
Background removal is the method in which the
isolation of the leaf disk in the photo is done. This
step is necessary because if the background has a
similar color to that of the mildew, then false positive
readings for mildew can occur. The steps for
background removal are shown in Figure 3 and the
respective outputs are shown in Figure 4.
Figure 3: Steps for background removal.
Figure 4: Background removed mirco- and macro- scopic
level photos.
2.3 HSV Masking
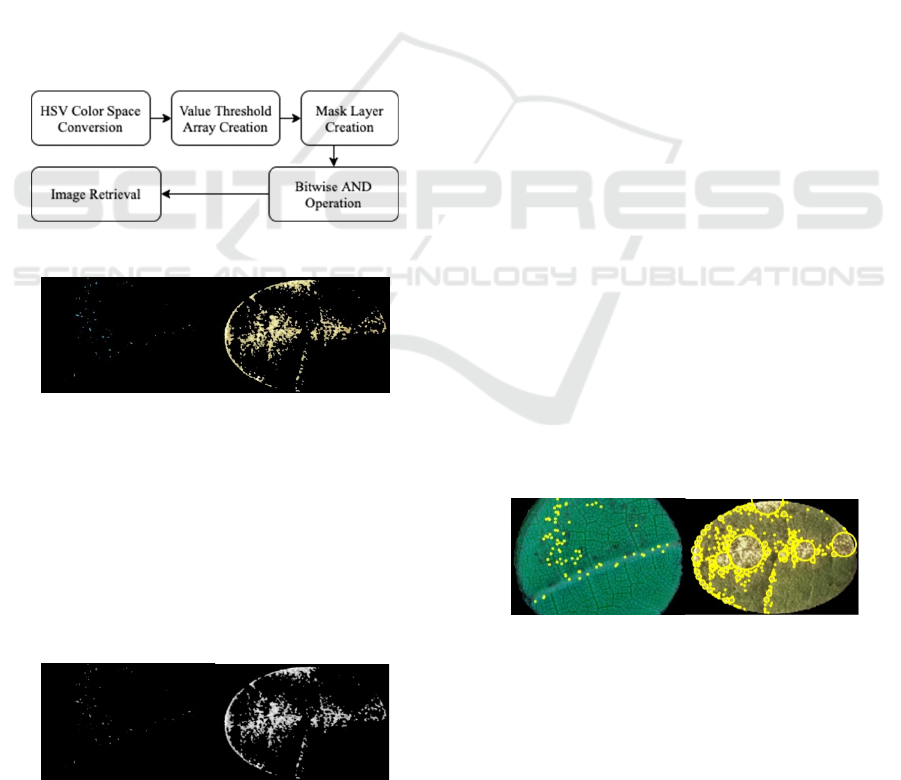
HSV masking is the phase where the areas within the
leaf disk that do not contain what is classified as
mildew are masked out prior to blob detection and
quantification. To accomplish this, an image’s color-
space must be converted from BGR to HSV. The set
of equations in (1) and (2) are used to convert 8/16-
bit images from BGR color space to HSV color space.
SIGMAP 2021 - 18th International Conference on Signal Processing and Multimedia Applications
82
Since BGR values can only lie between 0 and 255,
and HSV values lie between 0 and 255 for both
Saturation and Value and 0-360 for Hue, an extra step
of conversion is necessary to fit the Hue values
between 0 and 255 for 8-bit images.
V → max(R,G,B)
S → { V – min(R,G,B) if V ≠ 0
0 otherwise }
H → { 60(G-B)/(V – min(R,G,B)) if V = R
120 + 60(B-R)/ (V-min(R,G,B)) if V = G
240 + 60(R-G)/(V-min(R,G,B)) if V = B }
(1)
H → H /2
(2)
Since HSV is in three-dimensional space, two
threshold arrays or vectors are created to specify a
minimum and maximum threshold value. With this
threshold, utilizing the third number, namely the
value we want to mask over, allows us to keep only
pixels whose value surpasses that of the minimum
threshold. The steps to complete HSV masking are
shown in Figure 5 and the respective output is
presented in Figure 6.
Figure 5: HSV masking steps.
Figure 6: Output from HSV masking on micro- and
macroscopic-level photos respectively.
2.4 Blob Detection
Blob detection is where the quantification of the
mildew occurs. Once the masking has been
completed, the resulting image that contains what is
assumed to be mildew is once again converted to
grayscale. The grayscale images shown in Figure 7
are computed by equation (3).
Figure 7: Grayscale version of sample photos after HSV
masking.
Y → 0.299 · R + 0.587 · G + 0.114 · B
(3)
g(x,y,t) = (1/2πt) · e
-(x²+y²)/2t
(4)
L(x,y;t) = g(x,y,t) ꞏ f(x,y)
(5)
Once the grayscale image is returned, we then
utilize Blob detection for quantification of the
mildew. There are three types of Blob detection
algorithms that we initially considered - Laplacian of
Gaussian, Difference of Gaussians and Determinant
of Hessian. We chose to use the Laplacian of
Gaussian approach since it gave us the most accurate
and fastest output of the three. We capture the
different size blobs found throughout the image using
the equations in (4)-(7).
Equation (4) is used to convolve the input image,
which produces a third equation that dictates how one
function is shaped by another. Equation (5) shows the
scale space representation of the original image after
the convolution. The scale space representation is
essentially a representation of the photo, with a
Gaussian filter on it that amplifies as the scale t grows
in number. Then we apply the Laplacian Operator (6)
to (5) that determines the blobs of scale t as per
(Lindeberg, 2013).
▲²L = L
xx
+ L
yy
(6)
Following the application of (6) to (5), the
Laplacian of Gaussian algorithm outputs the
following:
detectedBlob = (x, y, σ)
(7)
Here, (x, y) are the coordinates of the blob and σ
is the standard deviation of the gaussian kernel which
detected the blob. Now, the radius of a single blob is
≈ √2 σ. At this point, we use equation (8) to obtain the
radius of the blob.
r = σ · √2
(8)
A = π · r²
(9)
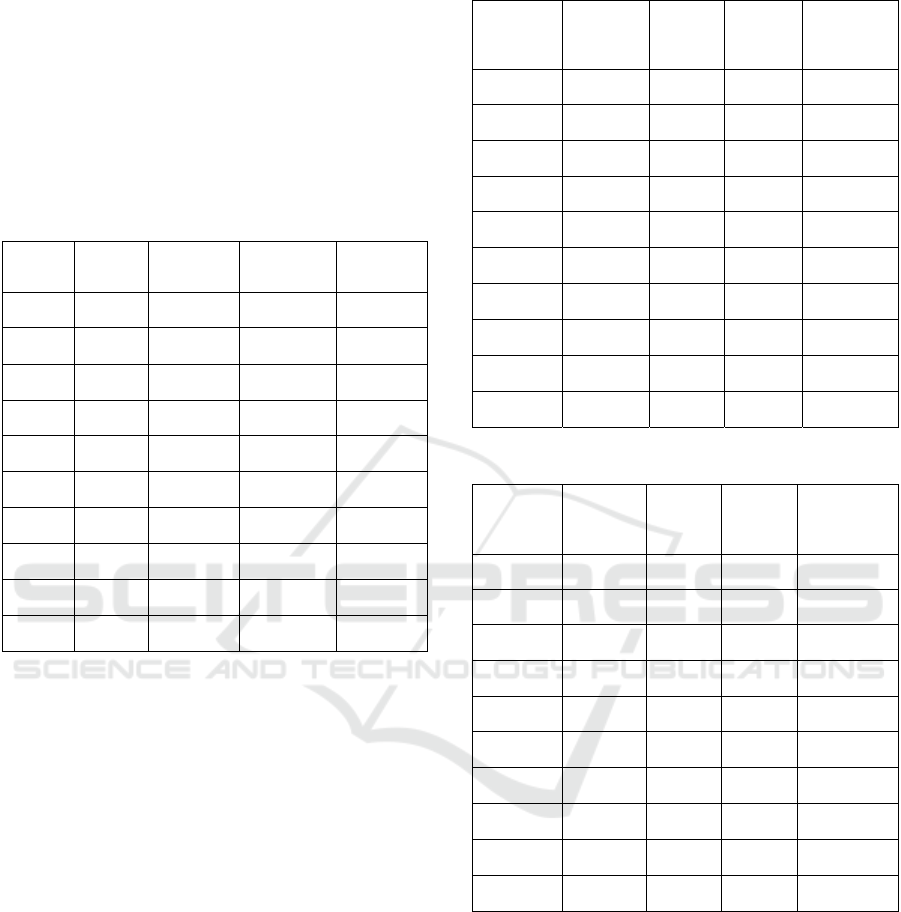
Figure 8: Detected blobs on micro- and macroscopic-level
photos.
Now that we have the radius of said blob at
coordinates (x,y), we find the area of a circle with said
radius found by using (9). We can draw circles for n
blobs that were detected in the photo, as shown in
Figure 8, and sum their areas together to retrieve the
total area taken up by the blobs on a single leaf disk
Towards Automatic Detection and Quantification of Mildew on Grape Leaf Disks
83
in pixel units. Once we retrieve all of the blob areas,
we then use equation (11) to compute a leaf to mildew
ratio.
TotalBlobArea = Σ A
i
(for i =0 to i=n)
(10)
TotalBlobArea/NonBlackPixelsAfterBR · 100
(11)
3 EVALUATION
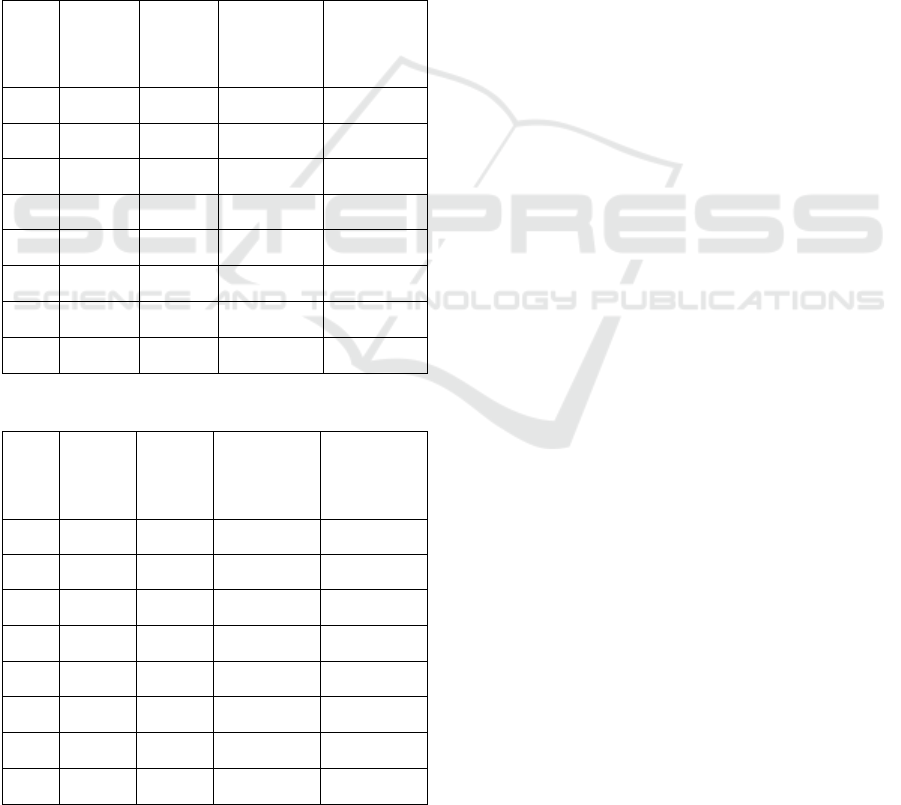
Table 1: Mildew growth over a 6-day period.
Tray
Number
Photo
Number
Mildew %
at 2dpi
Mildew % at
4dpi
Mildew %
at 6dpi
1 1 0.1193466 0.0 0.0055978
2 1 0.9503586 0.0413495 0.0117653
3 1 0.0061946 0.33576335 0.0369914
4 1 0.3733049 0.3357633 0.3462468
5 1 0.1799363 0.097927 0.1035048
6 1 0.3605445 0.4817453 0.0835825
7 1 0.0625684 0.0995543 0.092579
8 1 0.0550063 0.0791701 0.175554
1 2 1.6531951 1.3868999 2.173476
2 2 0.5955877 0.5151023 0.3470667
We tested our approach in two facets: growth tracking
and quantification/detection. The growth tracking
ability of our approach was tested by selecting a
singular plant and tracking its mildew growth. This
test was conducted on 10 of the 48 microscopic level
images. For the data presented in Table I, we tested
10 different samples over a six-day span. From the
reported results, we observed two trends, namely,
decreasing and increasing. However, the other trend
shown in the table, by rows 1 and 9, is that of
fluctuation. This trend is caused by the thresholding
aspect of the approach and the appearance of cell
structures that closely resemble the coloring of the
mildew. Also, the mildew requires viable cells to live
in, and therefore, if cells die then so does the mildew.
This phenomenon causes fluctuation in the amount of
mildew detected and quantified over the test period.
For quantification/detection evaluation, we tested
on two different computers - a laptop with 8GB of
RAM with an Intel Core i7 CPU @ 2.80GHZ
processor and a desktop with 32GB of RAM with an
Intel Core i7 CPU @ 3.2GHz processor. We will refer
to these two test machines as Machine A and Machine
B, respectively.
Table 2: Leaf to mildew ratios - Machine A.
Days Past
Inoculation
(DPI)
Photo Date
Tray
Number
Photo
Number
Mildew %
(pixels)
2 9-15-18 3 1 0.0
6 9-19-18 5 2 0.1533935
6 9-19-18 7 2 1.4873933
6 9-19-18 1 1 0.0059786
6 9-19-18 8 1 0.1755536
6 9-19-18 6 1 0.0835783
4 9-17-18 4 1 0.3357634
4 9-17-18 2 1 0.0413495
2 9-15-18 1 2 1.6531951
2 9-15-18 3 2 0.8965174
Table 3: Leaf to mildew ratios - Machine B.
Days Past
Inoculation
(DPI)
Photo Date
Tray
Number
Photo
Number
Mildew %
(pixels)
4 9-17-18 2 2 0.0059078
2 9-15-18 4 1 0.07149801
2 9-15-18 5 1 0.05584116
6 9-19-18 1 1 0.0
6 9-19-18 7 1 0.0555477
6 9-19-18 8 1 0.1271250
4 9-17-18 3 1 0.0651918
4 9-17-18 6 1 0.2470763
2 9-15-18 6 2 0.0609538
2 9-15-18 8 2 0.4067885
We tested the system with the following tests: 1
photo, 2 photos, 3 photos, 4 photos, up to 8 photos at
a time. Since we tested a total of 36 photos per test for
Tables II and III, we display only the first 10 results
without losing any important information. In Table II,
we showcase the quantification results from Machine
A, and in Table III, we showcase the results from
Machine B. Photos were chosen at random from three
date folders of 9-15-18, 9-17-18 and 9-19-18 and then
from one of the 8 tray folders within each date. The
photos selected for this test were also solely at the
microscopic level since the macroscopic-level photos
SIGMAP 2021 - 18th International Conference on Signal Processing and Multimedia Applications
84
did not belong to a specific date folder. The mildew
percentages are in terms of pixel which is calculated
by dividing the number of pixels that existed after
removing the background by the number of pixels the
blobs of mildew took up. Higher density of mildew
produces a higher mildew percentage overall. Since
these numbers are computer generated, they were
same for both machines, which is why we chose the
photos at random for both machines. Also, for the two
machines, there was a difference in the HSV mask
threshold values. The 8GB machine ran with a
threshold value of 170 and the 32GB machine ran
with a threshold of 180. This explains the difference
in the outputs of row 4, column 5 in both Tables II
and III.
Table 4: Detection-Quantification runtimes - Machine A.
# of
Tested
Photos
Avg.
Detection
Runtime
(ms)
Total
Detection
Runtime
(ms)
Avg.
Quantification
Runtime (ms)
Total
Quantification
Runtime (ms)
1 1.3819 1.3819 374.1365 374.1365
2 5.85395 11.70789 498.7908 997.5817
3 129.16577 387.4973 649.2774 1947.83229
4 104.20022 416.80089 976.5763 3906.30529
5 14.38234 71.91169 823.5583 4117.79159
6 40.49657 249.7939 958.8845 5753.307709
7 53.75931 376.151 1031.3986 7219.79089
8 60.03296 480.2636 1451.8371 11614.6974
Table 5: Detection-Quantification runtimes - Machine B.
# of
Tested
Photos
Avg.
Detection
Runtime
(ms)
Total
Detection
Runtime
(ms)
Avg.
Quantification
Runtime (ms)
Total
Quantification
Runtime (ms)
1
8.4423 8.4423 274.828 274.828
2
4.9833 9.9665 371.36 742.721
3
79.1446 237.4338 349.585 1048.750
4
68.0872 272.3490 646.065 2584.260
5
66.904 334.5221 1062.229 5311.147
6
59.927 359.5607 1151.012 6906.069
7
13.547 94.8268 1050.920 7356.4422
8
76.119 608.9588 1416.313 11330.505
Table IV and Table V showcase the detection-
quantification run-time performance for 8 different
tests. As expected, we see a trend in which as the
numbers increase, the total and average times
increase as well. However, depending on the amount
of quantification or detection time that one photo
takes depend on the amount of mildew that resides
within the photo itself. Again, as with the mildew
percentages found in Tables II and III, less resistant
plants will take a longer time to detect and quantify
the mildew that resides on the leaf disks.
4 CONCLUSIONS
In this paper, we proposed a multi-step approach for
the detection and quantification of mildew diseases
that reside on either the top or the bottom of grape
leaves. We are working towards a fully automated
detection process to perform quantitative analysis of
the mildew growth in an outdoor setting. It will add
value to the process of selectively breeding grapes
based on their resistance.
The approach presented in this paper contains the
ability to detect and quantify mildew at both the
microscopic and macroscopic level. To acclimate to
the issue of thresholding, we used a number between
170 to 180 to retrieve the most optimal results. Any
value less than 170 for the set of photos we used
allowed for more cell structures and anomalies within
the photo, e.g., lens flare from the camera and
microscope, to be captured and identified as mildew
as well. Therefore, we recommend that a threshold
number between 170 and 180 be used to eliminate
majority of the false positives that could occur during
detection.
We also observed that the utilization of a blur
while thresholding allowed for more accurate results.
Because of the intricacies of the cells, the blue helped
to soften some of the cell pixels that could produce
false positives. To accomplish this, a gaussian blur of
3 was applied on the photos and anything higher
causes pixels to become too blurry for the blob
detection algorithm to pick them up correctly.
In the future, we plan to continue adjusting
threshold values and begin testing the approach with
more than just grape leaf disks to determine if the
approach can successfully capture mildew diseases of
other kinds that may grow on other plants as well. An
example could be, looking at the powdery mildew,
known as Podosphaera leucotricha, that grows on
apples.
Towards Automatic Detection and Quantification of Mildew on Grape Leaf Disks
85

REFERENCES
Awate, A., Deshmankar, D., Amrutkar, G., Bagul, U., &
Sonavane, S. (2015). Fruit disease detection using
color, texture analysis and ANN. 2015 International
Conference on Green Computing and Internet of Things
(ICGCIoT), pp. 970-975.
Bierman, A., LaPlumm, T., Cadle-Davidson, L., Gadoury,
D., Martinez, D., Sapkota, S., & Rea, M. (2019). A
high-throughput phenotyping system using machine
vision to quantify severity of grapevine powdery
mildew. Plant Phenomics.
Cruz, J. A., Yin, X., Liu, X., Imran, S. M., Morris, D. D.,
Kramer, D. M., & Chen, J. (2016). Multi-modality
imagery database for plant phenotyping. Machine
Vision and Applications, 27(5), pp. 735-749.
Li, G., Ma, Z., & Wang, H. (2011). Image recognition of
grape downy mildew and grape powdery mildew based
on support vector machine. International Conference
on Computer and Computing Technologies in
Agriculture, pp. 151-162.
Lindeberg, T. (2013). Scale-space theory in computer
vision (Vol. 256). Springer Science & Business Media.
Sandika, B., Avil, S., Sanat, S., & Srinivasu, P. (2016).
Random forest based classification of diseases in grapes
from images captured in uncontrolled environments.
2016 IEEE 13th International Conference on Signal
Processing (ICSP), pp. 1775-1780.
Vijayakumar, J., & Arumugam, S. (2012). Recognition of
powdery mildew disease for betelvine plants using
digital image processing. International Journal of
Distributed and Parallel Systems, 3(2), 231.
SIGMAP 2021 - 18th International Conference on Signal Processing and Multimedia Applications
86
