
Prediction of G-protein Coupled Receptors using Deep Learning: A
Review
Anuj Singh, Arvind Kumar Tiwari
Department of Computer Science & Engineering, Kamla Nehru Institute of Technology, Sultanpur, Uttar Pradesh, India
Keywords: G-protein coupled receptors, glutamate, G proteins, signalling, amino acid composition, guanosine
triphosphate (GTP), guanosine diphosphate (GDP).
Abstract: The biggest super classes of the membrane proteins are G-protein coupled receptors as well as GPCRs are
very significant for drug design goals. GPCRs are sometimes known as heptahelical receptor as well as
seven-transmembrane receptor. GPCRs are accountable for several physicochemical and biological
activities like cellular growth, neurotransmission, smell as well as vision. This paper presents a review
related to current approaches to predict GPCRs. Extensive research on GPCRs have progressed to novel
discoveries that open undiscovered and promising drug design opportunities and efficient drug-targeting G-
protein coupled receptors therapies. This paper concentrates primarily on the process of deep learning to
estimate GPCRs.
1 INTRODUCTION
GPCRs are often addressed to the heptahelical
receptor or even the seven-transmembrane receptor.
A protein found in the cell membrane which also
binds extracellular substances as well as
communicates information to an intracellular
molecule named as a G protein. In cell membranes
of a large variety of species, like animals, crops,
microorganisms as well as invertebrates, GPCRs are
present in cell membranes of a large variety of
species, like animals, crops, microorganisms as well
as invertebrates(Dorsam, 2007). There are various
forms of G Protein-coupled receptor about 1,000
forms are identified by the human genome only.
Robert J. Lefkowitz, an American molecular
biologist, showed the presence of GPCRs in the year
1970s (Strader, 1994).
A wide class of proteins containing as even
transmembrane helical structural motif are GPCRs
(Karnik, 2003). G protein-coupled receptors are
huge number of related proteins which sense
molecules out of cell as well as initiate cellular
reactions. Coupling with G proteins GPCRs mostly
travel throughout the cell membrane seven times.
There seem to be two major pathways of signal
transduction concerning GPCRs one pathway is
cAMP signal and another one pathway is
phosphatidylinositol signal pathway (Gilman, 1987).
When a ligand binds to the GPCRs, it induces
conformational alterations in the G Protein-coupled
receptors that enable this one to serve as a transfer
mechanism for guanine nucleotides. GPCRs are a
big drug priority and about 34% of all licensed drugs
from the FDA target 108 elements of this group
(Hauser, 2018). Another rapidly emerging field of
pharmaceutical science is the long-discovered
relationship among G Protein-coupled receptors and
several endogenous as well as exogenous
compounds (Trzaskowski, 2012).G Protein-coupled
receptors convey extracellular signals through
membrane in plasma of intracellular effectors
through use of heterotrimeric G proteins (Pierce,
2002). GPCRs initiate drastic conformation
alterations disclosing intracellular sites which also
communicate with it as well as activate G proteins.
G proteins correspond to GTPase group which
consist of three subunits, α, β as well as γ, from
which β subunits as well as γ comprise the βγ-
subunits (Cabrera-Vera, 2003). This induces GDP
dissociation bound to a subunit of the Gα as well as
its substitution to the GTP. Gα-GTP along with Gβγ
subunit control downstream effectors (See Figure: 1)
(Harhammer, 1996).
Singh, A. and Tiwari, A.
Prediction of G-protein Coupled Receptors using Deep Learning: A Review.
DOI: 10.5220/0010563200003161
In Proceedings of the 3rd International Conference on Advanced Computing and Software Engineering (ICACSE 2021), pages 101-105
ISBN: 978-989-758-544-9
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
101
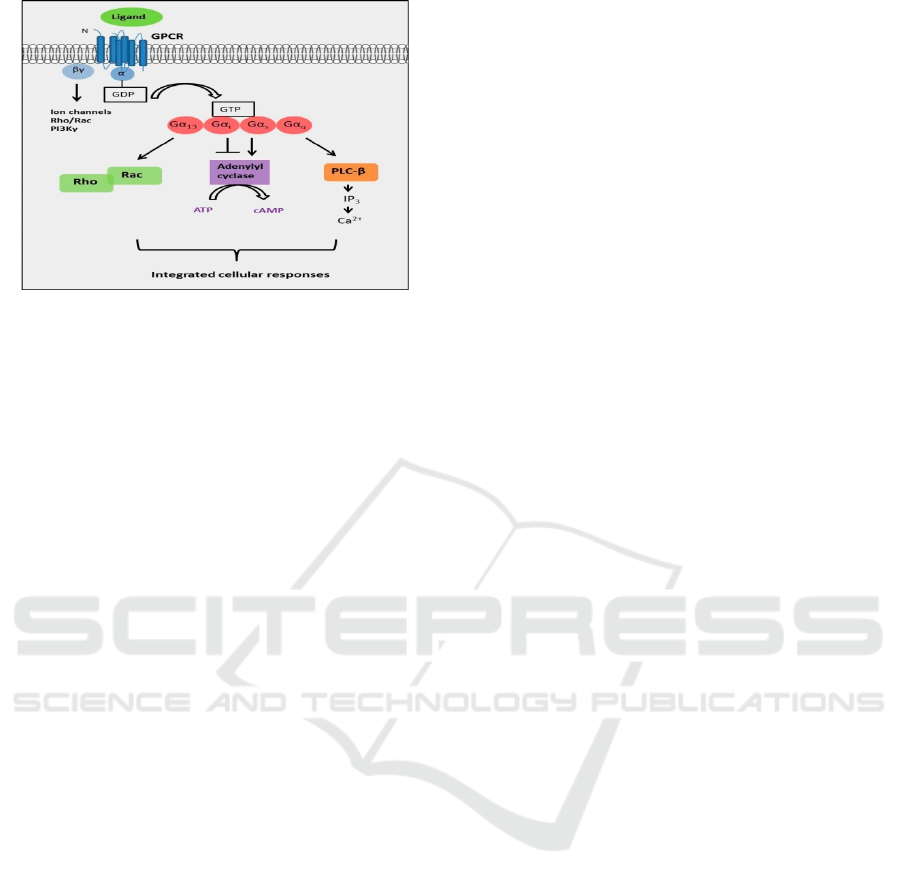
Figure1: G Signal of GPCRs through heterotrimeric G
proteins (Lynch, 2016).
2 RELATED WORKS
In paper (Bhasin, 2004), author used a SVM based
method, GPCRpred, for determination of G-protein
coupled receptor families as well as subfamilies
from dipeptide composition of proteins. When tested
with the help of 5-fold cross-validation, system
categorized GPCRs versus non-GPCRs with 99.5%
accuracy. In addition, the model can estimate 5 main
groups of GPCRs with an aggregate MCC of 0.81 as
well as efficiency of 97.5%. In paper (Liao, 2016),
the authors used physical-chemical properties,
coming from SVM-Prot, reflecting the G-protein
coupled receptors. They used random forest
classification technique to differentiate them from
all other sequences of proteins. In this work the
average classification accuracy was 91.61% as well
as 0.9282 was the average AUC.
G-protein-coupled receptors are main group of
receptors for the cell surface as well as one of the
most active sites of therapeutic drugs. Many of the
G-protein-coupled receptors functions are uncertain
along with identifying their ligands as well as signal
transduction pathways are time intensive and costly.
It prompts everyone meting a crucial problem: how
to develop an integrated method for the
classification of receptors coupled with G-proteins
family to assist in classification of medicines as well
as speed up the medicine research process. It is hard
to estimate the categorisation of G-protein-coupled
receptors by means of traditional sequence
alignment strategies due to their extremely divergent
nature. The covariant discriminant predictor was
implemented to estimate the families of G-protein-
coupled receptors in order to deal with such a
scenario (Chou, 2005). In paper (Rosenbaum, 2009),
the authors illustrated latest structural research of
high-resolution findings have been given into the
molecular pathways of activation as well as
constitutive function of G-protein coupled receptors.
Many of the physiological reactions to hormones,
neurotransmitters and stimulants in the atmosphere
are mediated by GPCRs and as therapeutic goals,
they have tremendous potential for a wide range of
infections. Our sense of hearing, our scent, our taste,
and our discomfort are mediated by GPCRs. They
are active in mechanisms of cell identification as
well as interaction, and have thus developed as an
influential superfamily for drug goals.
Unfortunately, for only one GPCR, the atomic-level
structure is open, making it hard to use structure-
based skills to solve medicines as well as mutation
research. They have currently developed approaches
of first principles for forecasting GPCR structure
and forecasting the sites of ligand binding and
relative binding allegiances (Vaidehi, 2002).
In paper (Basith, 2018), authors concentrated
primarily on the concepts of G-protein coupled
receptors drug development focused on
cheminformatics. They provide a complete analysis
of the cheminformatics methods focused on ligands
and structures, which are best demonstrated by
studies of GPCRs. In addition, an effective fusion of
ligand-based experience and structure-based
experience, like an incorporated solution, is also
addressed, which is appearing as a potential
cheminformatics-based G-protein coupled receptors
drug development technique. The current GPCR
structural biology development offers novel
visibility into ligand binding, conformational
dynamics, as well as signalling results regulation.
With the current techniques to multi-dimensional
drug action development, these insights allow
detailed classification of drugs along with their
pharmacodynamic features, which can be related to
the receptor structure as well as estimates of the
effectiveness of preclinical drugs (Wootten, 2018).
In paper (Popov, 2018), author developed a
comprehensive analytical method called
CompoMug, which uses Sequence-based research,
structural experience as well as a developed model
of machine learning to effectively forecast
stabilizing mutations in GPCRs. In paper (Carpenter,
2016), author illustrated the structure of a minimal G
protein, mini-Gs, which consist only from the
adenylate cyclase triggering G protein Gs of the
GTPase system. Mini-Gs are a thin, soluble protein
that, in absence of sub-units of Gβγ, effectively
combines GPCRs.
Extensive research on GPCRs have linked to
novel developments that open undiscovered and
ICACSE 2021 - International Conference on Advanced Computing and Software Engineering
102

promising drug design prospects and cost effective
drug-targeting GPCR therapies. This included the
development of unique signaling mechanisms like
ligand promiscuity occurring in cross-talks of
multitarget ligands, allosteric modulation and the
development of receptor homo as well as oligomers
that can be analyzed effectively with the help of
analytical modeling. Computer-aided approaches for
drug discovery can lower the price of creating drugs
by approximately 50% (Kaczor, 2016). In paper
(Bartuzi, 2017), the authors concentrated on
advances in docking of G protein-coupled receptors.
Appropriate statistical restoration of real ligand-
receptor method is known as molecular docking. In
paper (Schneider, 2018), the author illustrated latest
discoveries of hybrid coarse-grained membrane
protein approaches. They concentrate on in-house
molecular mechanics/coarse-grained approach. They
demonstrate that our molecular mechanics/coarse-
grained method is capable of capturing the atomistic
information of ligand binding interaction.
In paper (Vaidehi, 2016), authors identified the
existing structure of the analytical approaches that
provide inputs into G protein-coupled receptors
allosteric communication as well as explain how
allosteric modulators can be constructed with this
knowledge. The binding of ligands in the
extracellular region to GPCRs conveys the stimulus
to the intracellular region to activate coupling with
effector proteins. The method of this allosteric
contact appears to be mostly unexplored. In paper
(Foster, 2019), author reported the pairing of
cognate peptides as well as receptors. They define
common features that
reveal additional possible
peptidergic signaling mechanisms by
combining
selective genomics through 313 organisms and
bioinformatics over all protein sequences as well as
architectures of individual class A G protein-coupled
receptors. They fused 17 potential endogenous
ligands with five orphan G protein-coupled receptors
correlated with disorders involving developmental,
nervous as well as reproductive system diseases
employing three orthogonal biochemical assays.
In paper (Kobilka, 2007), the author illustrated
dynamic design of G-protein coupled receptors
activation structure along with functionality mainly
based on spectroscopic analyses of purified human
adrenergic β2 receptor. In paper (Shiraishi, 2019),
author established an original peptide descriptor-
incorporated SVM to estimate 22 pairs of
neuropeptides G protein-coupled receptors. For a 41
p% hit rate, the predicted pair signaling assays
identified 1 homologous neuropeptide and 11 Ciona-
specific neuropeptides G protein-coupled receptors.
In paper (Xiao, 2011), author developed a novel
Predictor by combining pseudo-amino acid
composition functional domainas well as the
pseudo-amino acid composition low-frequency
Fourier range. This novel predictor is named GPCR-
2L, in which "2L" implies a two-layer predictor.
GPCR-2L's total hit rate in recognizing there are
almost 97.2 percent of proteins as GPCRs or non-
GPCRs. In paper (Guo, 2006), author established a
novel technique ACC transform based support
vector machine to determine precision of coupling
among G protein-coupled receptors with G-proteins.
The main sequences of amino acids are converted
into vectors dependent on amino acids' key
physicochemical characteristics as well as the
content is converted by the implementation of ACC
transformation into a uniform matrix. Support vector
machine is qualified and tested through jackknife
testing for nonpromiscuous coupled G protein-
coupled receptors as well as promiscuous coupled G
protein-coupled receptors.
In paper (Chou, 2002), the author developed a
rapid system based on sequences to classify their
various G-protein-coupled receptors. In cellular
signaling networks that control different metabolic
functions like vision, odor, neurotransmission,
inflammation, cellular metabolism and cell
development, G-protein-coupled receptors perform a
major position of significance. For perception of
human physiology as well as sickness, such proteins
are really significant. Several pharmaceutical study
initiatives have tried to explain their composition
and purpose. These are hard to crystallize, and
therefore a few of them seem to not melt in
traditional solvents so very less number of GPCRs
structures have been identified. In paper (Wess,
1997), author outlined current evidence extracted
from structural, molecular genetics, biochemical, as
well as biophysical research that have cast fresh
insight on these processes and seek to combine
them.
3 CONCLUSIONS
The biggest super classes of the membrane proteins
are G-protein coupled receptors as well as GPCRs
are very significant for drug design goals. GPCRs
are sometimes known as heptahelical receptor as
well as seven-transmembrane receptor. GPCRs are
accountable for several physicochemical and
biological activities like cellular growth,
neurotransmission, smell as well as vision. This
paper has presented a review for current approaches
Prediction of G-protein Coupled Receptors using Deep Learning: A Review
103

to predict GPCRs. Extensive research on GPCRs
have progressed to novel discoveries that open
undiscovered and promising drug design
opportunities and efficient drug-targeting G-protein
coupled receptors therapies. This paper has
concentrated primarily on the process of deep
learning to estimate GPCRs.
REFERENCES
Dorsam, R. T., & Gutkind, J. S. (2007). G-protein-coupled
receptors and cancer. Nature reviews cancer, 7(2), 79-
94.
Strader, C. D., Fong, T. M., Tota, M. R., Underwood, D.,
& Dixon, R. A. (1994). Structure and function of G
protein-coupled receptors. Annual review of
biochemistry, 63(1), 101-132.
Karnik, S. S., Gogonea, C., Patil, S., Saad, Y., &
Takezako, T. (2003). Activation of G-protein-coupled
receptors: a common molecular mechanism. Trends in
Endocrinology & Metabolism, 14(9), 431-437.
Gilman, A. G. (1987). G proteins: transducers of receptor-
generated signals. Annual review of biochemistry,
56(1), 615-649.
Hauser, A. S., Chavali, S., Masuho, I., Jahn, L. J.,
Martemyanov, K. A., Gloriam, D. E., & Babu, M. M.
(2018). Pharmacogenomics of GPCR drug targets.
Cell, 172(1-2), 41-54.
Trzaskowski, B., Latek, D., Yuan, S., Ghoshdastider, U.,
Debinski, A., & Filipek, S. (2012). Action of
molecular switches in GPCRs-theoretical and
experimental studies. Current medicinal chemistry,
19(8), 1090-1109.
Pierce, K. L., Premont, R. T., & Lefkowitz, R. J. (2002).
Seven-transmembrane receptors. Nature reviews
Molecular cell biology, 3(9), 639-650.
Cabrera-Vera, T. M., Vanhauwe, J., Thomas, T. O.,
Medkova, M., Preininger, A., Mazzoni, M. R., &
Hamm, H. E. (2003). Insights into G protein structure,
function, and regulation. Endocrine reviews, 24(6),
765-781.
Harhammer, R., Gohla, A., & Schultz, G. (1996).
Interaction of G protein Gβγ dimers with small
GTP‐binding proteins of the Rho family. FEBS letters,
399(3), 211-214.
Lynch, J. R., & Wang, J. Y. (2016). G protein-coupled
receptor signaling in stem cells and cancer.
International journal of molecular sciences, 17(5), 707.
Bhasin, M., & Raghava, G. P. S. (2004). GPCRpred: an
SVM-based method for prediction of families and
subfamilies of G-protein coupled receptors. Nucleic
acids research, 32(suppl_2), W383-W389.
Liao, Z., Ju, Y., & Zou, Q. (2016). Prediction of G
protein-coupled receptors with SVM-prot features and
random forest. Scientifica, 2016.
Chou, K. C. (2005). Prediction of G-protein-coupled
receptor classes. Journal of proteome research, 4(4),
1413-1418.
Rosenbaum, D. M., Rasmussen, S. G., & Kobilka, B. K.
(2009). The structure and function of G-protein-
coupled receptors. Nature, 459(7245), 356-363.
Vaidehi, N., Floriano, W. B., Trabanino, R., Hall, S. E.,
Freddolino, P., Choi, E. J., ... & Goddard, W. A.
(2002). Prediction of structure and function of G
protein-coupled receptors. Proceedings of the National
Academy of Sciences, 99(20), 12622-12627.
Basith, S., Cui, M., Macalino, S. J., Park, J., Clavio, N. A.,
Kang, S., & Choi, S. (2018). Exploring G protein-
coupled receptors (GPCRs) ligand space via
cheminformatics approaches: impact on rational drug
design. Frontiers in pharmacology, 9, 128.
Wootten, D., Christopoulos, A., Marti-Solano, M., Babu,
M. M., & Sexton, P. M. (2018). Mechanisms of
signalling and biased agonism in G protein-coupled
receptors. Nature reviews Molecular cell biology,
19(10), 638-653.
Popov, P., Peng, Y., Shen, L., Stevens, R. C., Cherezov,
V., Liu, Z. J., & Katritch, V. (2018). Computational
design of thermostabilizing point mutations for G
protein-coupled receptors. Elife, 7, e34729.
Carpenter, B., & Tate, C. G. (2016). Engineering a
minimal G protein to facilitate crystallisation of G
protein-coupled receptors in their active conformation.
Protein Engineering, Design and Selection, 29(12),
583-594.
Kaczor, A. A., Rutkowska, E., Bartuzi, D., Targowska-
Duda, K. M., Matosiuk, D., & Selent, J. (2016).
Computational methods for studying G protein-
coupled receptors (GPCRs). Methods in cell biology,
132, 359-399.
Bartuzi, D., Kaczor, A. A., Targowska-Duda, K. M., &
Matosiuk, D. (2017). Recent advances and
applications of molecular docking to G protein-
coupled receptors. Molecules, 22(2), 340.
Schneider, J., Korshunova, K., Musiani, F., Alfonso-
Prieto, M., Giorgetti, A., & Carloni, P. (2018).
Predicting ligand binding poses for low-resolution
membrane protein models: Perspectives from
multiscale simulations. Biochemical and biophysical
research communications, 498(2), 366-374.
Vaidehi, N., & Bhattacharya, S. (2016). Allosteric
communication pipelines in G-protein-coupled
receptors. Current opinion in pharmacology, 30, 76-
83.
Foster, S. R., Hauser, A. S., Vedel, L., Strachan, R. T.,
Huang, X. P., Gavin, A. C., ... & Gloriam, D. E.
(2019). Discovery of human signaling systems: pairing
peptides to G protein-coupled receptors. Cell, 179(4),
895-908.
Kobilka, B. K., & Deupi, X. (2007). Conformational
complexity of G-protein-coupled receptors. Trends in
pharmacological sciences, 28(8), 397-406.
Shiraishi, A., Okuda, T., Miyasaka, N., Osugi, T., Okuno,
Y., Inoue, J., & Satake, H. (2019). Repertoires of G
protein-coupled receptors for Ciona-specific
neuropeptides. Proceedings of the National Academy
of Sciences, 116(16), 7847-7856.
ICACSE 2021 - International Conference on Advanced Computing and Software Engineering
104

Xiao, X., Wang, P., & Chou, K. C. (2011). GPCR-2L:
predicting G protein-coupled receptors and their types
by hybridizing two different modes of pseudo amino
acid compositions. Molecular Biosystems, 7(3), 911-
919.
Guo, Y., Li, M., Lu, M., Wen, Z., & Huang, Z. (2006).
Predicting G‐protein coupled receptors–G‐protein
coupling specificity based on autocross‐covariance
transform. Proteins: structure, function, and
bioinformatics, 65(1), 55-60.
Chou, K. C., & Elrod, D. W. (2002). Bioinformatical
analysis of G-protein-coupled receptors. Journal of
proteome research, 1(5), 429-433.
Wess, J. (1997). G‐protein‐coupled receptors: molecular
mechanisms involved in receptor activation and
selectivity of G‐protein recognition. The FASEB
Journal, 11(5), 346-354.
Prediction of G-protein Coupled Receptors using Deep Learning: A Review
105
