
A Blind Noise Estimation and Removal in Histopathological Images
Shiksha Singh, Rajesh Kumar
Department of Electronics and Communication, J K Institute of Applied Physics and Technology, University of Allahabad,
Prayagraj, Uttar Pradesh, India
Keywords: Histopathology images, Noise Estimation, Image Processing
Abstract: With the advancement in technology for digital pathology, a huge chunk of the visual dataset is prepared for
medical experts for disease diagnosis and grading. The introduction of noise in various image modalities in
the medical field can distress the result of diagnosis which could lead to inappropriate disease grading and
hence delay in treatment. In this, a blind noise estimation and removal technique is proposed for
histopathology images. The model uses the wavelet transformed image and block selection approach with a
block size of eight for noise estimation. The noise estimated in the model is Gaussian, Poisson, and speckle.
The proposed approach is verified on images of Breakhis dataset with all four-magnification scale. The
performance of the proposed approach is shown through parameter Signal-to-noise ratio (SNR), mean
square error (MSE), root mean square error (RMSE) and peak signal to noise ratio (PSNR)
1 INTRODUCTION
Noise in the digital image is defined as inappropriate
and unwanted information residing in an image. This
unwanted noise may affect the image quality in
different ways. The disturbance is created in pixel
values of the image, and hence there is some random
variation in pixel value. In medical image analysis,
the estimation of noise variance is of utmost
importance. To check the stability and working
performance of the detection model for disease
diagnosis, noise recognition, as well as elimination,
is important. With the improvement in digital
pathology, a large amount of visual dataset is
generated which is available for computer-aided
diagnosis (CAD). The availability of dataset
encourages the researchers to develop CAD tools for
analysing the scanned images developed through
digital pathology. Based on the result obtained from
CAD tools, the grading of the disease is performed.
For a precise and accurate diagnosis of medical
images, it is very essential that the image taken
through different technique remains free from blur,
noise, and artefacts. The different image acquisition
techniques end up in accumulating a large number of
numbers of the pixel in per unit area to flourish
high-resolution image. They thrive to capture high
quality leads to noise accumulation in the resultant
image. These noises mask the essential feature
which leads to incorrect grading of the disease.
Table 1 below reports a brief overview of different
image modalities along with noise present in such
images. The table below gives the image capturing
developing techniques related to different medical
image modalities. Due to different faults in different
capturing process noise are introduced in the image.
There are various noise models for different medical
images, and it is shown in Table 1.
There are models developed for other image
modalities such as MRI, X-Ray, Ultra-sound, CT etc.
using different approaches such as filtering,
statistical, block selection, noise variance etc. It can
be observed from the literature that no such model is
developed for histopathology images. This paper
proposes a blind noise estimation model for
histopathology images. There is no prior knowledge
regarding the noise model hence we are using blind
noise estimation approach. In this mode model, the
image is transformed into the wavelet domain and
perform block division of image in the continuous
diagonal, vertical and horizontal component. For
each block median absolute deviation is calculated.
For denoising, the noisy level and estimated noise
level is differentiated. Due to the importance of
luminance in the microscopic image the Color space
used is YCbCr.
Singh, S. and Kumar, R.
A Blind Noise Estimation and Removal in Histopathological Images.
DOI: 10.5220/0010562700003161
In Proceedings of the 3rd International Conference on Advanced Computing and Software Engineering (ICACSE 2021), pages 67-72
ISBN: 978-989-758-544-9
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
67
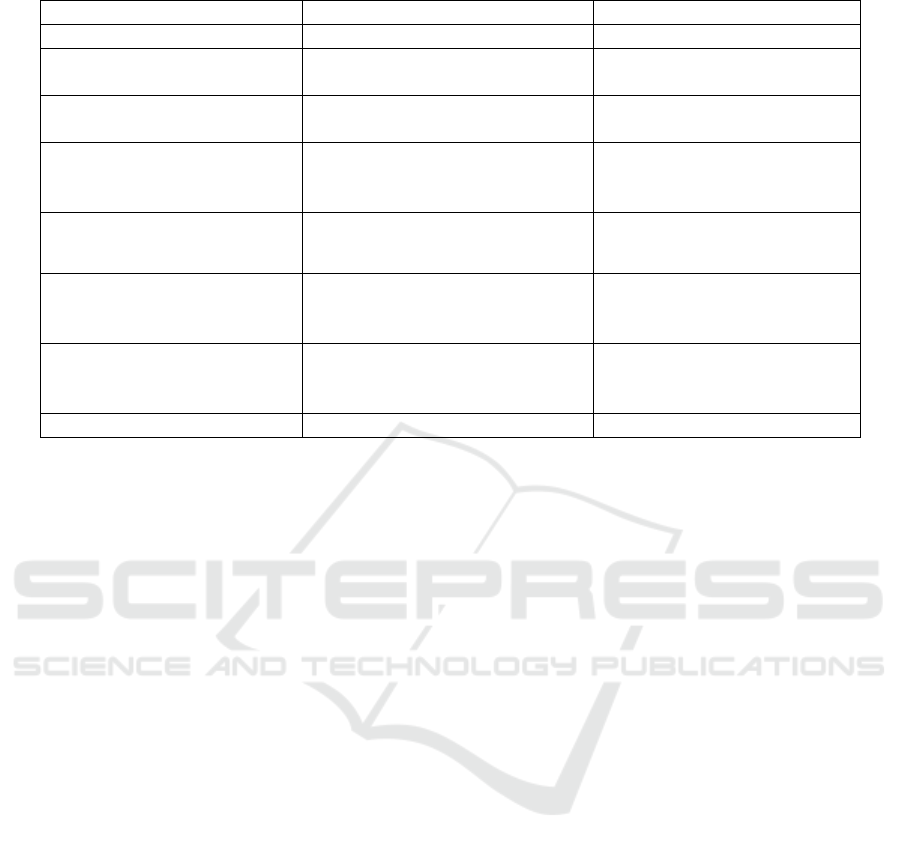
Table 1: Image modalities with noise and acquiring technique
The paper is organized as: already existing
model of noise estimation are given in “Related
work”. The proposed approach and the dataset used
to validate the model is explained in “Material and
Method.” This section gives a detailed discussion
regarding noise estimation, noise model and Discrete
wavelet transform.
The experimental work and result analysis are
discussed in “Experimental Result”. The future
direction and work proposed is conclude in section
“Conclusion”.
2 RELATED WORKS
To best of our knowledge, no such noise estimation
model for histopathology images has been sated in
the literature. So, we have reviewed paper based on
other medical image modality for noise estimation.
There are different types of image-modality in the
medical image. Every image has a different
acquisition technique based on those image
capturing approaches; different type of noise is
introduced in a different image. The table
summarizes the image capturing technique along
with the noise present in that image (Goyal, Dogra,
Agrawal, & Sohi, 2018)(Dogra, Goyal, Agrawal, &
Sohi, 2017). There are several noise models and
noise estimation approaches are reported till date
such as statistical approaches, patch-based, filter-
based and block selection based (Ram & Choudhary,
2014)(Kaur, 2015).
Pieere Gravel et.al. (2004) has developed a
method for analysing the statistical property for
analysing the statistical property of noise. The model
developed establishes the association between the
intensity of the image and variance of image. The
proposed model was examined on MRI and X-Ray
images with Gaussian, Poisson as well as Rician
noise(Gravel, Beaudoin, & De Guise, 2004). M.N.
Nobi et.al. (2010) has developed noise reduction
model for MRI and ultra-sound images having
Rician noise and speckle noise. The model integrates
the median filter and mean filter(Yousuf & Nobi,
2010). Pierrick Coupe et.al. (2010) has presented an
object-based model to estimate Rician noise in MRI
using median absolute deviation(Coupé et al., 2010).
GnanambalIlango et.al. (2011) has proposed a
hybrid approach of noise estimation using different
filtering techniques. The estimation technique is
used on brain tumour image for Gaussian noise
removal (Ilango & Marudhachalam, 2011). Xuvyu
Pan et.al. (2012), the authors have presented a blind
noise estimation model for CT images. Contrast
band filters are used for estimating the noise and for
denoising PCA with local pixel grouping is used
(Pan, Zhang, & Lyu, 2012). Jose V. Manjon et. al.
(2015) has given a two-step approach for Rician
noise estimation in MRI. The proposed approach
involves the filtering of the noisy image using no-
local principal component analysis(PCA) and then
using a filtered image as a guide for the non-local
mean filter (Manjón, Coupé, & Buades, 2015). F.F.
Ting et.al. (2016) has proposed a rapid noise
variance estimation method for magnetic resonance
Ima
g
e Modalit
y
Technique T
y
pe of Noise
X-Ra
y
X-ra
y
p
ro
j
ection Gaussian & Poisson Noise
Computed-Tomography (CT)
Cross-sectional body x-ray
p
ro
j
ection
Gaussian & Quantum Noise
Positron Emission
Tomo
g
raph
y
(PET)
Radioactive tracing Gaussian Noise
Single-photon emission
Computed Tomography
(SPECT)
Picturization performed through
the nuclear substance.
(Gamma camera)
Gaussian Noise
Magnetic -Resonance
Imaging (MRI)
Transition in the energy of the
photon
Gaussian, Richian, and
Rayleigh noise
Ultra-sound
Reflection of the temporal wave
with
hi
g
h frequenc
y
Gaussian & multiplicative
noise
Microscopic Biopsy
Tissue examined under a
microscope.
with H&E stains on i
t
Gaussian, Poisson, and
Multiplicative
Mammo
g
raph
y
Low dose x-ra
y
s
y
ste
m
Gaussian & Poisson Noise
ICACSE 2021 - International Conference on Advanced Computing and Software Engineering
68

image and computed tomography. The author has
used Gabor Wavelet Laplacian convolution (GWLC)
for noise variance estimation. The type of noise
discussed in the work is Rician noise (Ting, Sim, &
Wong, 2017). Rajesh Kumar et.al. (2017) has
proposed an approach for segmentation of
microscopic images for cancer grading. The
proposed approach segments the cell and nuclei in
the existence of Poisson noise. The authors have
used the partial differential equation of order four
which relies on the non-linear filter for noise
estimation (Kumar et al., 2017). Table.2. gives a
brief over of the noise estimation model discussed
by various researchers and noise model they have
considered.
Table 2: Brief overview noise estimation model stated in the literature
3 METHOD AND MATERIALS
In the proposed approached we have used block
selection method for noise estimation. Fig.2. shows
a block diagram of the proposed architecture. Since
it is blind noise estimation technique introduction of
noises such as Gaussian, Poisson, and speckle, are
made in images of Breakhis dataset. For validating
our approach, we have used benchmark dataset for
experimental work. For noise estimation and
denoising we have used images from BreakHis
dataset. System configuration with 2 GB GPU, 8 GB
Ram i5 processor has been used. The Matlab version
2017b is used for performing an experiment. The
images in the dataset are in RGB colour space they
are transformed into YCbCr colour space and
performed wavelet transform. This section is
subdivided into sections- 3.1) Dataset 3.2) Pre-
processing 3.3) Noise model 3.4) Wavelet transform
3.5) Noise estimation 3.6) Performance measure.
3.1 Dataset
Breast Cancer Histopathology Database (BreakHis)
is publicly available dataset and is prominently used
for breast cancer detection. The images are
developed by staining the tissue collected through
surgical open biopsy and staining them with H&E. A
total of 7909 images are them out of which 2440 are
benign and 5429 are malignant and they are of
magnification factor 40X, 100X, 200X and 400X.
For validation of proposed architecture, a single
image from each magnification level is
taken(Spanhol, Oliveira, Petitjean, & Heutte,
2016)(“Breast Cancer Histopathological Database
Reference Image Modality Type of Noise Approaches
(Gravel et al.,
2004)
MRI
X-Ray
Gaussian, Poisson,
&
Rician
Relationship between noise variance
and image intensity
(Yousuf & Nobi,
2010)
MRI
Ultra-soun
d
Rician& Speckle
Integration of median filter an
d
Mean filter.
(Coupé et al.,
2010)
MRI Rician
Two-step approach involves filtering
image using non-local PCA and then
filtered image used as a guide for
non-local
Mean filter.
(Ilango &
Marudhachalam,
2011)
MRI
Gaussian, Sal
t
-
pepper &
Speckle
Hybrid filter through Topological
approach
(Manjón et al.,
2015)
CT, MRI
Additive white
gaussian
N
oise (AWGN)
Contrast band filter
(Ting et al., 2017) MRI, CT Rician
Gabor Wavelet Laplacian
Convolution
(Kumar,
Srivastava, &
Srivastava, 2017)
Microscopic
Biopsy
Poisson
Fourth-order partial differential
equation based on non-linear filter
A Blind Noise Estimation and Removal in Histopathological Images
69

(BreakHis),” 2014). Fig.1. shows the images used in
the proposed approach:
Ma
g
nification Facto
r
40x 100x 200x 400x
Figure 1: Images from BreakHis Dataset
3.2 Pre-processing
The luminance of the coloured images is of utmost
importance. And in case of histopathological
images, the Color stain signifies the characteristics
of an image. So, keeping this in mind we have
chosen YCbCr for conversion. The conversion of the
RGB image to colour image is:
Y Cb Cr
R G B
0.299 0.168 0.499
0.587 0.331 0.418
0.114 0.500 0.081
(1)
Figure 2: Proposed Architecture for noise estimation and removal
3.3 Noise Model
During digital image acquiring of slides, various
sensors are coupled with the microscope. This led to
noise introduction in an image due to a decrease in
contrast of tissue structure. This occurs due to the
lack of proper light and long duration of exposure.
Due to this there is a scarcity of photon in sensor and
hence the shifting electrons inside the chip get lost
and noise intrudes the image. Noise is characterized
as a random variable since it is simply a fluctuation
in pixel value. The random variables have some
probability distribution, which links it with statistical
values which is the probability of occurrence (Kaur,
2015). The basic assumption regarding noise nx,y
noises is an additive random signal that is white
Gaussian noise with zero mean value and noise is of
high frequency. The noise in an image J
x, y
is
represented as
J
x, y
J
x, y
nx,y (2)
Where, J
is the degraded image.
In the proposed approach we have introduced
Gaussian, Poisson, and speckle noise in images for
noise estimation and denoising purpose.
3.3.1 Gaussian Noise
It is a statistical noise which has the probability
density function (PDF) equals to the normal
distribution (Gonzalez & Woods, 2002). It can be
mathematically given as:
I
Ix,yG
(3)
Where I x, y is the noiseless image and G
is
Gaussian PDF it is given as:
G
=
√
e
, ∞𝑎∞ (4)
Here a represents the intensity, a
is
average(mean) of intensity, and σ standard
deviation.
3.3.2 Poisson Noise
The Poisson is introduced in the image due to
random fluctuation in photons from source ray
emission. This result in temporal and spatial
randomness(Gonzalez & Woods, 2002). The PDF
for Poisson Noise is given as
P
a
!
e
(5)
Here n represents the total number of pixel and p
shows the ratio between the noise pixel to the total
number of pixels.
3.3.3 Speckle Noise
Another noise which is very common and can be
present in microscopic images are speckle
noise(Gonzalez & Woods, 2002). Speckle noise is
most common multiplicative noise in medical
images. It is represented as
Color Conversion
(RGB to YCbCr)
Discrete
Wavelet
Transform
Noise
Estimation
Denoising
ICACSE 2021 - International Conference on Advanced Computing and Software Engineering
70

J
J
x, y
J
x, y
S
(6)
Where S
shows a random noise with zero mean
Gaussian PDF.
3.4 Wavelet Transform
The wavelet signifies the analysis and representation
of multiresolution images (Jaiswal & Srivastava,
2020) Wavelet transformation are most frequently
used in edge detection (Jiang, Shen, Jiang, & Lam,
2009) and image denoising(Coifman&Donoho,
1995). At low frequency, the wavelet transform
gives high resolution and with high frequency it
gives high resolution time. We can get better noise
estimation of an image in the wavelet domain. So,
image is transformed in wavelet domain. This
transformation is applied over YCbCr image.
Let us consider Ψx as wavelet of 1-D signal,
then the scaling parameter p and shifting parameter r
is given as:
Ψ
,
x
Ψ
(7)
Where, f(x) 1-D signal and its wavelet transform is
given as:
T
,
p, r
f
x
Ψ
,
xdx (8)
3.5 Noise Estimation
Upon excluding the edge, the noise estimation is
performed by block division into continuous
diagonal, vertical, and horizontal component and
then the noise statics are calculated for each block
obtained. The noise statistics calculated for blocks
are mean absolute deviation. The block size here is
taken as eight. The denoising is performed by
differentiating noisy image and estimated noise of an
image.
3.6 Performance Measure
The quality of the image needs to be quantified. The
metrics are put under category object fidelity and
subjective fidelity. For testing the performance of
enhancement approach, we have calculated error and
signal to noise ratio. MSE is error calculated
between the input image and the processed image.
SNR is the ratio between signal amplitude and noise
amplitude(Jain, 1989). The unit of SNR is dB. The
formula for calculation of MSE, RMSE SNR, and
PSNR is given as
E
∑∑
J
x, y
J′x,y
(9)
RMSE
√
E
(10)
Here l and m are dimensions of input image
J(x,y) and J’(x,y) is the processed image
SNR 20 log 10
(11)
PSNR 20 log 10
(12)
Here S
is signal amplitude and N
is noise
amplitude.
4 EXPERIMENTAL RESULT
The model explained in this paper is validate on the
images taken from BreakHis dataset. The proposed
approach is tested on images with different
magnification scale and varying noise model. The
experiment is performed by selecting five random
images from the database and then their value is
represented in Table.3 Then the SNR, PSNR, MSE,
and MSE is calculated value is computed to quantify
the image quality given by the estimation model.
Lower the value of MSE, RMSE and higher value of
SNR, PSNR shows the betterment of enhancement
procedure. Table 3. Gives a brief overview of SNR,
PSNR, MSE and RMSE values corresponding to
different noise and magnification scale. It is
observed for the table that magnification of image
does not affect the signal amplitude and noise
amplitude ratio.
Table 3: SNR value of the proposed model on the different
magnification factor
Image
Type
Type of
Noise
MSE RMSE SNR PSNR
40X Speckle 0.3586 0.5968 49.4785 4.5128
Gaussian 0.4649 0.6805 48.5876 3.3607
Poisson 0.4663 0.6846 48.1698 3.3197
100X Speckle 0.3932 0.6250 49.2943 4.1031
Gaussian 0.4677 0.6826 48.4031 3.3309
Poisson 0.4904 0.6992 48.2628 3.1208
200X Speckle 0.3952 0.6273 49.2936 4.0695
Gaussian 0.4648 0.6804 48.5888 3.3619
Poisson 0.5182 0.7185 48.1157 2.8869
400X Speckle 0.4432 0.6632 48.2968 3.5933
Gaussian 0.5218 0.7441 47.2101 2.5686
Poisson 0.5734 0.7551 47.1749 2.5686
A Blind Noise Estimation and Removal in Histopathological Images
71

5 CONCLUSIONS
The noiseless image is every essential medical
domain; the detection accuracy totally relies on the
eminence of the image. As the work reported in
literature there are noise detection and removal
model developed for other modalities of the medical
image like X-Ray, MRI, Ultra-sound, CT, etc., but
there is no such model available for the microscopic
image. The model introduced in the paper estimates
the noise in microscopic image with assuming some
distributed noise such as Gaussian, Poisson, and
speckle. The approach is based on the blind noise
estimation technique using the block selection
method. The block size of the model is 8, DWT is
used because it accurately analyses the images with
abrupt changes as it is well localized in terms of
frequency and time. The denoising is performed
using differentiating estimated noise from noisy
image. The result is described in signal to noise ratio
and error is also calculated and the model performs
well for all the magnification level. The lower values
of MSE and RMSE and higher values of SNR &
PSNR indicates the betterment of proposed
enhancement model. In future we would like to
develop an estimation model based on the filtering
approach and for denoising statistical approach, this
could result in better SNR value.
REFERENCES
Breast Cancer Histopathological Database (BreakHis).
(2014). Retrieved September 30, 2019, from
https://web.inf.ufpr.br/vri/databases/breast-cancer-
histopathological-database-breakhis/
Coifman, R. R., & Donoho, D. L. (1995). Translation-
Invariant De-Noising.
Coupé, P., Manjón, J. V., Gedamu, E., Arnold, D., Robles,
M., & Collins, D. L. (2010). Robust Rician noise
estimation for MR images. Medical Image Analysis,
14(4), 483–493.
https://doi.org/10.1016/j.media.2010.03.001
Dogra, A., Goyal, B., Agrawal, S., & Sohi, B. S. (2017).
Anatomical and Functional Imaging Modalities : A
Brief Review, 9028, 113–118.
Gonzalez, R., & Woods, R. (2002). Digital image
processing. Prentice Hall.
https://doi.org/10.1016/0734-189X(90)90171-Q
Goyal, B., Dogra, A., Agrawal, S., & Sohi, B. S. (2018).
Noise issues prevailing in various types of medical
images. Biomedical and Pharmacology Journal, 11(3),
1227–1237. https://doi.org/10.13005/bpj/1484
Gravel, P., Beaudoin, G., & De Guise, J. A. (2004). A
method for modeling noise in medical images. IEEE
Transactions on Medical Imaging, 23(10), 1221–1232.
https://doi.org/10.1109/TMI.2004.832656
Ilango, G., & Marudhachalam, R. (2011). New hybrid
filtering techniques for removal of gaussian noise from
medical images. ARPN Journal of Engineering and
Applied Sciences, 6(2), 8–12.
Jain, A. (1989). Fundamentals of digital image processing.
Retrieved from
http://www.amazon.co.uk/Fundamentals-Processing-
Prentice-Information-
Sciences/dp/0133361659%5Cnhttp://dl.acm.org/citatio
n.cfm?id=59921
Jaiswal, A. K., & Srivastava, R. (2020). Time-efficient
spliced image analysis using higher-order statistics.
Machine Vision and Applications, 31(7–8).
https://doi.org/10.1007/s00138-020-01107-z
Jiang, W., Shen, T. Z., Jiang, W., & Lam, K. M. (2009).
Efficient Edge Detection Using Simplified Gabor
Wavelets. IEEE Transactions on Systems, Man, and
Cybernetics, Part B: Cybernetics, 39(4), 1036–1047.
https://doi.org/10.1109/TSMCB.2008.2011646
Kaur, S. (2015). Noise Types and Various Removal
Techniques. Nternational Journal of Advanced
Research in Electroni Cs and Communication
Engineering (IJARECE), 4(2), 226–230.
Kumar, R., Srivastava, S., & Srivastava, R. (2017). A
fourth order PDE based fuzzy c- means approach for
segmentation of microscopic biopsy images in
presence of Poisson noise for cancer detection.
Computer Methods and Programs in Biomedicine,
146, 59–68.
https://doi.org/10.1016/j.cmpb.2017.05.003
Manjón, J. V., Coupé, P., & Buades, A. (2015). MRI noise
estimation and denoising using non-local PCA.
Medical Image Analysis, 22(1), 35–47.
https://doi.org/10.1016/j.media.2015.01.004
Pan, X., Zhang, X., & Lyu, S. (2012). Blind local noise
estimation for medical images reconstructed from
rapid acquisition. Medical Imaging 2012: Image
Processing, 8314, 83143R.
https://doi.org/10.1117/12.910857
Ram, B. P., & Choudhary, S. (2014). Survey Paper on
Different Approaches for Noise Level Estimation and
Denoising of an Image. International Journal of
Science and Research, 3(4), 618–622.
Spanhol, F. A., Oliveira, L. S., Petitjean, C., & Heutte, L.
(2016). A Dataset for Breast Cancer Histopathological
Image Classification. IEEE Transactions on
Biomedical Engineering, 63(7), 1455–1462.
https://doi.org/10.1109/TBME.2015.2496264
Ting, F. F., Sim, K. S., & Wong, E. K. (2017). A rapid
medical image noise variance estimation method.
Proceedings of 2016 International Conference on
Robotics, Automation and Sciences, ICORAS 2016.
https://doi.org/10.1109/ICORAS.2016.7872628
Yousuf, M. A., & Nobi, M. N. (2010). A New Method to
Remove Noise in Magnetic Resonance and Ultrasound
Images. Journal of Scientific Research, 3(1), 81.
https://doi.org/10.3329/jsr.v3i1.5544
ICACSE 2021 - International Conference on Advanced Computing and Software Engineering
72
