
Biomedical Dataset Recommendation
Xu Wang, Frank van Harmelen and Zhisheng Huang
Vrije University Amsterdam, De Boelelaan 1105, 1081 HV Amsterdam, The Netherlands
Keywords:
Dataset Recommendation, Scientific Datasets.
Abstract:
Dataset search is a special application of information retrieval, which aims to help scientists with finding the
datasets they want. Current dataset search engines are query-driven, which implies that the results are limited
by the ability of the user to formulate the appropriate query. In this paper we aim to solve this limitation by
framing dataset search as a recommendation task: given a dataset by the user, the search engine recommends
similar datasets. We solve this dataset recommendation task using a similarity approach. We provide a simple
benchmark task to evaluate different approaches for this dataset recommendation task. We also evaluate the
recommendation task with several similarity approaches in the biomedical domain. We benchmark 8 different
similarity metrics between datasets, including both ontology-based techniques and techniques from machine
learning. Our results show that the task of recommending scientific datasets based on meta-data as it occurs
in realistic dataset collections is a hard task. None of the ontology-based methods manage to perform well on
this task, and are outscored by the majority of the machine-learning methods. Of these ML methods only one
of the approaches performs reasonably well, and even then only reaches 70% accuracy.
1 INTRODUCTION
Dataset search is an application of dataset re-
trieval, which is specialization of information re-
trieval (Kunze and Auer, 2013). Dataset search aims
to help people to find datasets they want. Scien-
tists, journalists and decision makers would be typi-
cal users of such a dataset search service. A dataset
is “a set of related observations which are organized
and formatted for a specific purpose” (Chapman et al.,
2020). A dataset can be of widely different for-
mats: tables, files, images, structured objects or oth-
ers (Google Developers, 2021).
As one particular application case of dataset
search, a dataset search engine returns datasets based
on a user query. A number of such dataset search
engines are now operating in practice, for exam-
ple Google Dataset Search
1
or the Mendeley Data
search engine
2
operated by Elsevier. Such query-
driven search engines come with the limitation that
the search engine is highly reliant on an appropriate
search query presented by the user.
In (Wang et al., 2020), we provided a recom-
mendation paradigm for dataset search, which can be
paraphrased as "if you like this dataset, you’ll also
1
https://datasetsearch.research.google.com/
2
https://data.mendeley.com/
like that one". This recommendation paradigm gives
us the motivation for this paper, with bringing a new
paradigm "if user like a query or a dataset, the dataset
search engine could recommend a similar dataset to
the user". This paradigm could be applied to a dataset
search engine in two scenarios: a) when users can
provide a search query with an appropriate descrip-
tion, the engine could recommend datasets similar to
the search query; b) when user cannot provide that
query, the user can provide a dataset as a query, and
the search engine will recommend a similar dataset.
In this paper we are aiming to answer following
research questions: 1) How to precisely define the
task of recommending similar datasets from queries
or from other datasets? 2) How to evaluate different
approaches to this recommendation task? 3) Can we
set up experiments to test this evaluation task in a spe-
cific scientific domain?
There are many related works on dataset recom-
mendation. In (Leme et al., 2013), they provide a
dataset recommendation task by using the link be-
tween dataset and bibliographic domain (e.g. DBLP).
In (Ellefi et al., 2016), dataset recommendation works
by considering schema overlap between datasets. In
(Patra et al., 2020), they provide a dataset recommen-
dation approach with help of publications and inter-
ests of researchers. In (Singhal et al., 2013), the au-
192
Wang, X., van Harmelen, F. and Huang, Z.
Biomedical Dataset Recommendation.
DOI: 10.5220/0010521801920199
In Proceedings of the 10th International Conference on Data Science, Technology and Applications (DATA 2021), pages 192-199
ISBN: 978-989-758-521-0
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
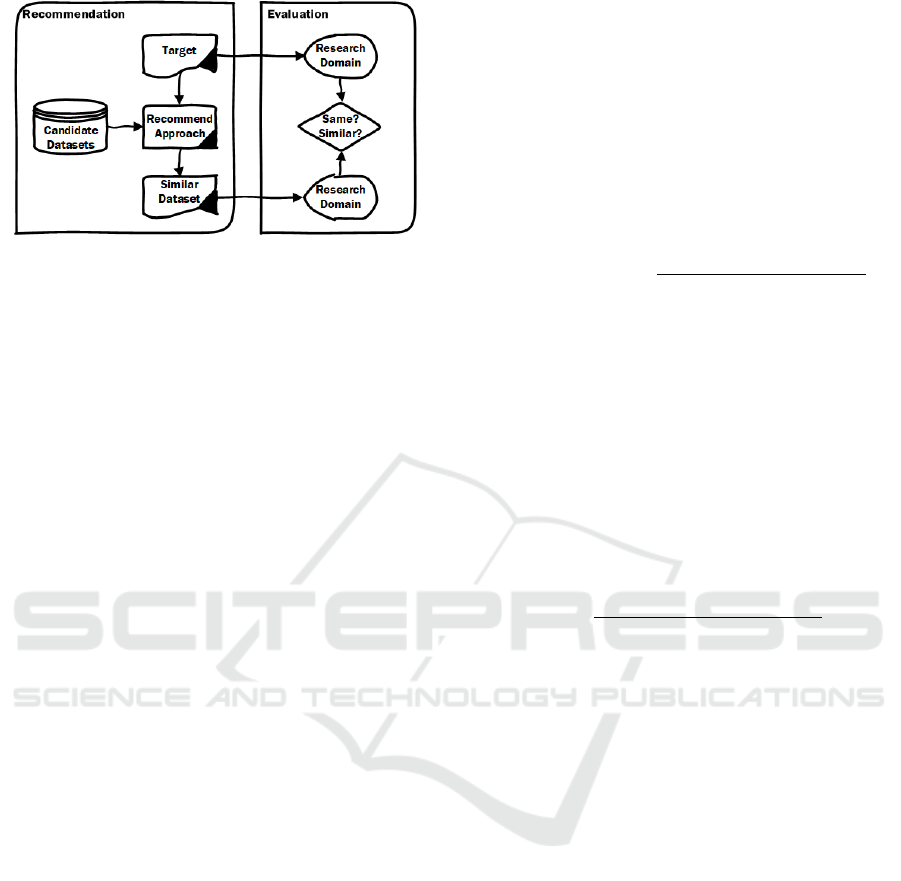
Figure 1: Overview of main works in this paper.
thors used the context of user’s interest to find inter-
esting research datasets. All these approaches dif-
fer from the approach we take in this paper, namely
providing dataset recommendations based on only the
meta-data descriptions of datasets as provided by the
authors of the datasets.
We given an overview of our paper in Figure 1,
which shows the two parts of our work: the recom-
mendation task and the evaluation of different ap-
proaches for this task. The recommendation task will
try to recommend similar datasets chosen from can-
didate datasets for a given target (which could be
a dataset or a query). The evaluation will evaluate
the recommendation work by checking if the recom-
mended dataset belongs to the same scientific do-
main as the target. This choice of success-criterion
(do we manage to recommend a dataset from the
same domain as the target) may sound like a low-
barrier. However, our experimental results in section
5 will actually show that even this seemingly simple
success-criterion is far from trivial to achieve.
The main contributions of this paper are: 1) We
provide a novel dataset recommendation task. This
task recommends similar datasets from a set of can-
didate datasets given a target that consists of an-
other dataset or a query. We also provide several
approaches for this similar dataset recommendation
task, by applying a number of similarity approaches.
2) We provide a procedure for evaluating the differ-
ent approaches to the similar dataset recommendation
task. Different from expert judgement, our (simpler)
evaluation task is to compare the research domain be-
tween the given dataset/query (the target) and the rec-
ommended dataset (the output). As argued above,
this seemingly easy success-criterion will turn out to
be already very hard to achieve by many of the ap-
proaches we test. 3) We run experiments to test sev-
eral approaches to the similar dataset recommenda-
tion task for targets in the biomedical domain. The re-
sults show that single approach (BM25) outperforms
others in our experiments.
2 PRELIMINARIES
In this section we will introduce the different similar-
ity definitions we will use in this paper.
Wu-Palmer. Wu-Palmer is an edge-based semantic
similarity metric to calculate the similarity between
concepts in hierarchical structure or ontology (Wu
and Palmer, 1994). The Wu-Palmer similarity be-
tween two concept C
A
and C
B
is defined as
Sim
WuP
(C
A
,C
B
) =
2 ∗ Path(R, LCS(C
A
,C
B
))
Path(R,C
A
) + Path(R,C
B
)
where LCS(C
A
,C
B
) is the least common subsumer of
C
A
and C
B
; Path(R, n) is the length of the path be-
tween the ROOT node R and node n in the ontology
structure.
Resnik. Resnik is a content-based semantic similar-
ity metric to calculate the similarity between concepts
with the help of their information content (Resnik,
1995). The Resnik similarity between two concepts
C
A
and C
B
is defined as:
Sim
Res
(C
A
,C
B
) = − log p(LCS(C
A
,C
B
))
= − log
∑
n∈words(LCS(C
A
,C
B
))
count(n)
N
where LCS(C
A
,C
B
) is the least common subsumer
of C
A
and C
B
; p(LCS(C
A
,C
B
)) is the probability of
LCS(C
A
,C
B
) in the definition of information content;
words(LCS(C
A
,C
B
)) is the set of concepts which are
subsumed by LCS(C
A
,C
B
) in the ontology structure;
N is the total number of concepts in the ontology
structure.
LDA. LDA (latent Dirichlet allocation) is a gener-
ative probabilistic model for a corpus, which could
used a set of topics to represent a document (Blei
et al., 2003). The LDA similarity between two con-
tent fields is calculated by first extracting topics from
each content field and then to check the coverage be-
tween two sets of topics.
Doc2Vec. Doc2vec is a method to create a numeric
representation of a document (Le and Mikolov, 2014).
The Doc2vec similarity metric between two content
fields is calculated by first transferring each of con-
tent fields into pretrained a vector space and then cal-
culate the distance between the two content fields in
this vector space.
Biomedical Dataset Recommendation
193

BM25. BM25 (Best Match 25) is a ranking func-
tion that scores documents based on their relevance to
a given text query (Robertson et al., 1995). The BM25
approach is used by search engines to rank matching
documents according to their relevance to a search
query. In this paper, we consider textual content of
our target as "query" and use BM25 to find relevant
content from candidate datasets.
Bert. Bert (Bidirectional Encoder Representations
from Transformers) is a multi-layer neural language
representation model (Devlin et al., 2019). We use
Bert to compute the similarity between two contents
fields by first transferring each content to a vector us-
ing Bert’s encoder. Then we will calculate the simi-
larity between the two vectors with cosine similarity.
This similarity score will be used as the similarity be-
tween two content fields.
3 DATASET RECOMMENDATION
In this section we will introduce the dataset rec-
ommendation task and a number of different ap-
proaches to this task based on similarity metrics be-
tween datasets.
3.1 Concept-based Similarity between
Datasets
Based on the similarity metrics we introduced above,
here we will introduce the similarity approach be-
tween datasets. Our concept-based similarity ap-
proach between datasets only focuses on the ontol-
ogy concepts extracted from the textual content of the
meta-data fields of datasets. These extracted ontology
concepts are the concepts which appear both in the
ontology and in the textual content of the meta-data.
Restricting the meta-data fields to the concepts
from ontologies will enable the use of ontology-based
similarity approaches. In particular, if the ontology
captures the terminology of a research domain, the
concept extraction of a dataset will contain the con-
cepts from that domain, and will ignore the "noise"
terms in the meta-data fields of datasets, which are
useless for similarity approach. For example, if we
use the UMLS ontology
3
for concept extraction, the
extracted concepts will always belong to the biomed-
ical research domain, and concepts from other do-
mains (e.g. Computer Science) will be ignored. This
will then benefit us when we generate recommenda-
tions for biomedical datasets.
3
https://www.nlm.nih.gov/research/umls/index.html
After the introduction of our concept extraction
approach, we will now introduce concept-based sim-
ilarity between datasets based on the Wu-Palmer and
Resnik metrics.
Def. 1 (Concept-based Similarity between Datasets).
Given two datasets D
A
and D
B
, and given a simi-
larity approach between concepts, denoted as sim.
Concept-based similarity ConSim(D
A
, D
B
) between
D
A
and D
B
is defined as follows:
ConSim(D
A
, D
B
) =
∑
{sim(c
A
, c
B
)|c
A
∈ D
A
, c
B
∈ D
B
}
|D
A
| ∗ |D
B
|
where c
A
is a concept from textual meta-data con-
tent of dataset D
A
and similarly c
B
; sim(c
A
, c
B
) is the
similarity between c
A
and c
B
, calculated by similarity
metric sim; |D
A
| is the number of all concepts in the
meta-data of D
A
and similarly |D
B
|.
We can instantiate this generic definition with spe-
cific similarity metrics by replacing sim with Wu-
Palmer or Resnik.
We also define concept-based similarity between a
query and a dataset. In this definition, we extract con-
cepts from the query and then apply the same concept-
based similarity calculation as above. The definition
of concept-based similarity between a query Q and a
dataset D is then as follows:
ConSim(Q, D) =
∑
{sim(c
Q
, c
D
)|c
Q
∈ Q, c
D
∈ D}
|Q| ∗ |D|
(1)
where c
Q
is a concept from content query Q and |Q|
is the number of all concepts occurring in Q.
3.2 Content-based Similarity between
Datasets
Here we will introduce the content-based similarity
between datasets which could be used with ML-based
and IR-based approaches. The content we consider
here is still the textual meta-data content of datasets,
which are title and description.
Def. 2 (Content-based Similarity between Datasets).
Given two datasets D
A
and D
B
. Given a similarity
approach between contents, denoted as sim. Con-
tent similarity ContSim(D
A
, D
B
) between D
A
and D
B
is the similarity between C
A
and C
B
, where C
A
is the
textual content of D
A
and C
B
is the textual content of
D
B
.
We can then instantiate this definition of content-
based similarity with content metrics such as LDA,
Doc2vec, BM25 and Bert. For LDA, BM25 and Bert,
we can directly apply them to content-based similar-
ity because all of them are similarity metrics between
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
194

contents. For Doc2vec, we consider the whole tex-
tual meta-data content of a dataset as a "document"
and then calculate the similarity between such "docu-
ments" of datasets.
Similar to concept-based similarity between query
and dataset, we also have to define content-based sim-
ilarity between query and dataset. We define content-
based similarity between a query Q and a dataset D as
followa:
ContSim(Q, D) = sim(C
Q
,C
D
) (2)
where C
Q
is the content of query Q; C
D
is the textual
meta-data content of dataset D; sim(C
Q
,C
D
) is a sim-
ilarity metric between contents, which could be LDA,
Doc2vec, BM25 and Bert.
3.3 The Similar Dataset
Recommendation Task
After introducing the two types of approaches for
the similarity between datasets (concept-based and
content-based), we will now introduce the task of rec-
ommending a similar dataset.
Def. 3 (Similar Dataset Recommendation Task SDR
for Dataset). Given a target dataset D
T
and a list
of candidate datsasets CL
D
, the similar dataset rec-
ommendation SDR task is to return a sorted list of
candidate datasets SCL
D
= {D
1
, D
2
, ......, D
n
} so that
Sim(D
T
, D
1
) ≥ Sim(D
T
, D
2
) ≥ ...... ≥ Sim(D
T
, D
n
) >
0, where D
1
, D
2
, ......, D
n
∈ CL
D
. Then D
1
∈ SCL
D
is
the dataset recommended by this task for D
T
, denoted
as RD
SDR
D
T
.
We also define the similar dataset recommenda-
tion task for a query.
Def. 4 (Similar Dataset Recommendation Task SDR
for a Query). Given a target query Q and a list
of candidate datasets CL
D
, the similar dataset rec-
ommendation task SDR is to return a sorted list of
candidate datasets SCL
D
= {D
1
, D
2
, ......, D
n
} so that
Sim(Q, D
1
) ≥ Sim(Q, D
2
) ≥ ...... ≥ Sim(Q, D
n
) > 0,
where D
1
, D
2
, ......, D
n
∈ CL
D
. Then D
1
in SCL
D
is
the dataset recommended by this task for Q, denoted
as RD
SDR
Q
.
After introducing the similar dataset recommen-
dation task for both dataset and query, we can now
turn to a variety of similar dataset recommendation
approaches by applying different similarity mentrics
to this task.
Also, we identify three types of recommendation
approaches, Concept-based and Content-based, and
Hybrid one. The Concept-based similar dataset rec-
ommendation approach applies concept-based simi-
larity metrics (Wu-Palmer or Resnik) to the dataset
recommendation task. The Content-based approach
on the other hand applies content-based similarity
metrics (LDA, Doc2vec, BM25 or Bert) to the dataset
recommendation task. Finally, the Hybrid approach
combines concept-based and LDA metrics.
4 EVALUATION FOR DATASET
RECOMMENDATION
In this section we will introduce both our evaluation
method and the gold standard we will use in the eval-
uation.
We deliberately choose an evaluation task that is
at first sign very easy to achieve: we count a dataset
recommendation as correct, if the recommendation is
in the same scientific domain as the original target. As
we will see, even this apparently rather easy evalua-
tion task will turn out to be difficult to achieve by al-
most all of the methods that we test in our experiment.
A benefit of this choice of evaluation task is that a
clear gold standard is available: In many online open-
source dataset repositories, such as figshare
4
or Har-
vard Dataverse
5
, there are categories or subjects that
show the subject-area or research domain and topics
of datasets. Two examples are shown in Figure 2 and
Figure 3.
Since in our evaluation procedure we will take all
our targets from biomedical datasets or queries, the
evaluation task comes down to predicting which of
the candidate datasets (some 200.000 datasets from
all kinds of scientific domains) belong to the biomed-
ical domain.
We can now give a more precise definition of our
evaluation task:
Def. 5 (Simple Evaluation Task on Similar Dataset
Recommendation for Dataset). Given a list of tar-
get datasets L
D
T
; a list of datasets L
D
C
as rec-
ommendation candidate datasets; a list of similar
dataset recommendation approaches L
SDR
; and the
gold standard scientific domains Standard
D
0
T
for each
D
0
T
in L
D
T
. The evaluation task for similarity dataset
recommendation is to find the approach SDR
0
from
L
SDR
so that for every SDR
00
∈ L
SDR
(SDR
00
6= SDR
0
),
|{RD
SDR
0
D
0
T
⊆
Domain
Standard
D
0
T
}| ≥ |{RD
SDR
00
D
0
T
⊆
Domain
Standard
D
0
T
}|, where ⊆
Domain
means subset relation-
ship on domain area; RD
SDR
0
D
0
T
⊆
Domain
Standard
D
0
T
means the domain of dataset RD
SDR
0
D
0
T
is the subset of
Standard
D
0
T
.
4
https://figshare.com/
5
https://dataverse.harvard.edu/
Biomedical Dataset Recommendation
195

Figure 2: Gold Standard Example 1 (figshare).
Figure 3: Gold Standard Example 2 (Harvard Dataverse).
and similarly for evaluating recommendations based
on a target that is a query:
Def. 6 (Simple Evaluation Task on Similar Dataset
Recommendation for Query). Given a list of target
datasets L
Q
; a list of datasets L
D
as recommenda-
tion candidate datasets; a list of similar dataset rec-
ommendation approaches L
SDR
, and the gold stan-
dard scientific domains Standard
Q
0
for each Q
0
in
L
Q
. The valuation task for similarity dataset rec-
ommendation is to find the approach SDR
0
from
L
SDR
so that for every SDR
00
∈ L
SDR
(SDR
00
6= SDR
0
),
|{RD
SDR
0
Q
0
⊆
Domain
Standard
Q
0
}| ≥ |{RD
SDR
00
Q
0
⊆
Domain
Standard
Q
0
}|, where ⊆
Domain
means subset relation-
ship on domain area; RD
SDR
0
Q
0
⊆
Domain
Standard
Q
0
means the domain of dataset RD
SDR
0
Q
0
is the subset of
Standard
Q
0
.
In all our evaluation matching between two do-
mains, we also consider synonyms of the domain.
This means that we do not only do exact matching,
but also match one domain to the synonym of that do-
main.
5 EXPERIMENTS
In this section we will introduce our experiments.
5.1 Preparation
First off, we will introduce the datasets and the gold
standard used in our experiments.
Target Datasets. Thanks to our colleagues from El-
sevier, all the datasets we used are from the Else-
vier data repository
6
. All the target datasets in our
experiments are 190 datasets derived from Elsevier
6
https://data.mendeley.com/research-data/
?repositoryType=NON_ARTICLE_BASED_
REPOSITORY
DataSearch
7
based on 19 biomedical queries (shown
in Table 1), with the top10 datasets per query. These
queries all come from a carefully validated set of test
queries for Elsevier’s Mendeley data search engine.
As a result, we can be certain that all of these 190
datasets are indeed from the biomedical domain, as
they are the top10 results on 19 validated biomedical
queries.
Candidate Datasets. We use 209.998 datasets from
Mendeley Data
8
. These datasets are randomly se-
lected from about 18 million datasets in the Elsevier
data repository. Recommendation datasets in our ex-
periments must be selected from these ≈210k candi-
date datasets. There are about 600 domains for these
candidate datasets without considering synonym of
domain.
These datasets contain textual meta-data (title and
description of the dataset), topic meta-data (scientific
domain of the dataset) and so on. In the recommen-
dation approach, we only consider the textual meta-
data (title and description) of datasets. We also use
the "topic" meta-data, (the "subjectArea" in the meta-
data schema of the datasets) only in the evaluation.
This is because of the particularity of our evalua-
tion approach. Our evaluation approach is to evalu-
ate the scientific research domain. In the metadata of
the dataset, the "topic" meta-data is the scientific re-
search domain. Therefore, we only consider topic in
the evaluation approach, not in the recommendation
approach.
Training Datasets. For training the ML-based dis-
tance metrics described above, we used ≈21M
datasets (21.110.372 to be precise) as our training cor-
pus. We obviously took care that this training set is
disjoint from the 210k candidate datasets, so as to
avoid data leakage from the training phase in our ex-
periment.
7
https://datasearch.elsevier.com/
8
https://data.mendeley.com/
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
196
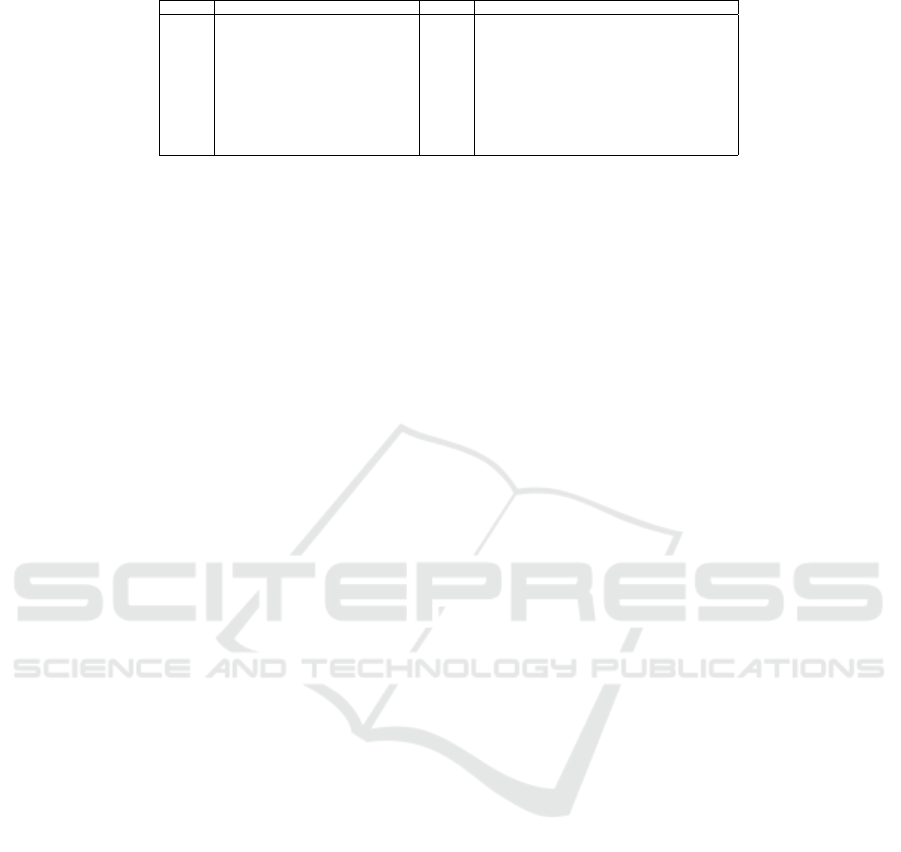
Table 1: The corpus of 19 biomedical queries.
Query Content Query Content
E2 Protein Degradation mechanisms E54 glutamate alcohol interaction
E7 oxidative stress ischemic stroke E66 calcium signalling in stem cells
E8 middle cerebral artery occlusion mice E67 phylogeny cryptosporidium
E17 Risk factors for combat PTSD E68 HPV vaccine efficacy and safety
E26 mab melting temperature E78 c elegans neuron degeneration
E28 mutational analysis cervical cancer E79 mri liver fibrosis
E31 metformin pharmacokinetics E80 Yersinia ruckeri enteric red mouth disease
E35 prostate cancer DNA methylation E89 Electrocardiogram variability OR ECG variability
E50 EZH2 in breast cancer E94 pinealectomy circadian rhythm
E88 Flavonoids cardiotoxicity
Gold Standard. According to what we discussed in
last section, we consider the categories or subjects of
datasets as gold standard for evaluation task. In the
≈210k candidate datasets, there are 60.484 datasets
which are labeled by "None" subject or "Uncatego-
rized".
Evaluation. We use check if domain of recom-
mended dataset is same or similar to the one of target
datasets by hand. This is doable and easy to do be-
cause it’s not hard to check if two domains are same
or similar for human (not even be a domain expert).
5.2 Experimental Set-up
In this part we will introduce the set-up of our two
evaluation experiments. In total we use eight dataset
recommendation approaches for these experiments:
• Two concept-based recommendation (with Wu-
Palmer and Resnik)
• Two hybrid recommendation (with LDA+Wu-
Palmer and LDA+Resnik)
• Four content-based recommendation (with LDA,
Doc2vec, BM25 and Bert_Base)
Exp1: Evaluation of Dataset Recommendation
Approaches from Dataset Targets. In this exper-
iment, we will evaluate the dataset recommendation
approaches when given a dataset as the input tar-
get. Given a list of recommendation approaches, the
pipeline of Exp1 is:
1. Using 19 biomedical queries to get 190 target
candidates: We send each query to the Elsevier
Datasearch API, and we select the top 10 returned
datasets as target candidates.
2. For each recommendation approach SDR in the
list of recommendation approaches given above:
(a) For each target dataset, using recommenda-
tion approach SDR to find the Top1, Top5 and
Top10 similar datasets from ≈210k candidate
datasets.
(b) We consider a dataset recommended by SDR as
a success-count when the domain of this dataset
meets the domain of the target dataset.
3. The recommendation approach with the best per-
formance is the one with the highest success-
count over all 190 target candidates.
Exp2: Evaluation of Dataset Recommendation
Approaches from Query Targets. Different from
Exp1, we will here recommend similar datasets from
a given query as target. Given a list of recommenda-
tion approaches, the pipeline of Exp2 is:
1. Using the 19 biomedical queries as target queries.
2. For each recommendation approach SDR from the
list of approaches given above:
(a) For each target query, we use SDR to find the
Top1, Top5 and Top10 similar datasets from
≈210k candidate datasets.
(b) We consider a dataset recommended by SDR as
a success-count when the domain of this similar
dataset meets the domain of the target query.
3. The recommendation approach with best perfor-
mance is the one with highest success-count over
19 target queries.
5.3 Results and Analysis
In this section we will show the results of the two ex-
perimental scenarios that we set up above. In Table 2
and Table 3, we show the results of two experimental
scenarios for both Exp1 and Exp2:
• Scenario 1: consider all TopN datasets, whether
or not they contained subject-area metadata.
• Scenario 2: consider only TopN datasets with
subject-area metadata.
Scenario 1. In the first experimental scenario
(shown in Table 2), we consider all the TopN simi-
lar datasets for each target dataset or query, even if
some similar datasets do not contain the subject-area
in their meta-data. In such a case, we take the conser-
vative position, and consider it a failure if the recom-
mendation approach recommends a dataset that has
Biomedical Dataset Recommendation
197
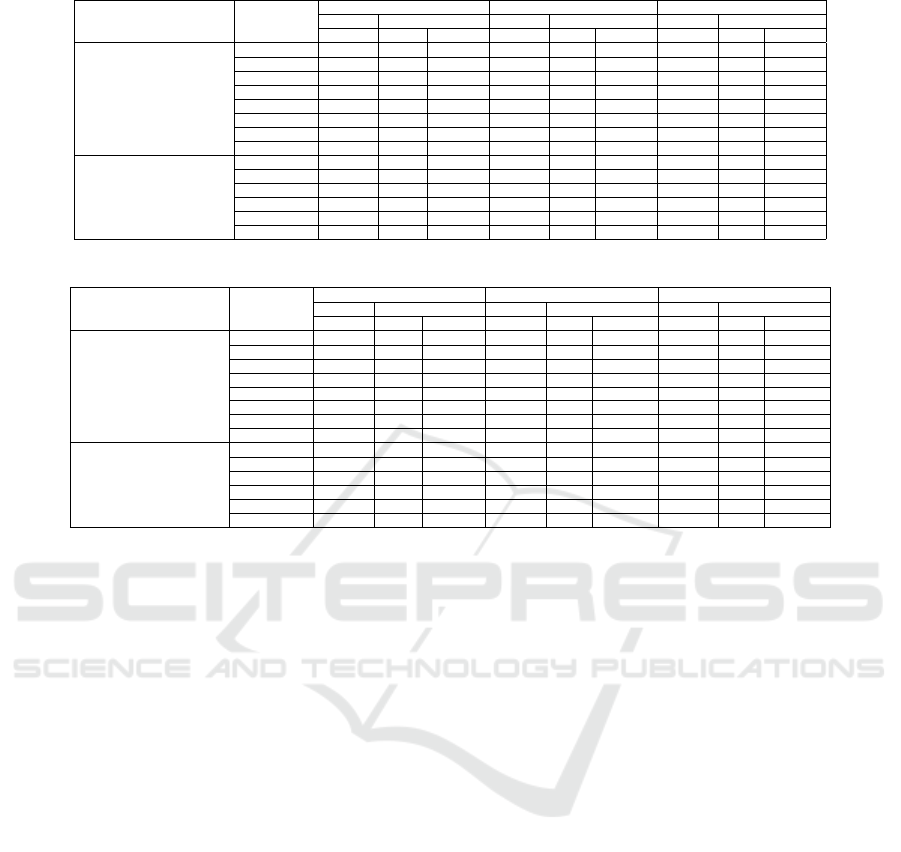
Table 2: Scenario 1: Top10, Top5 and Top1 results.
Experiment Measure
Top10 Judgement Top5 Judgement Top1 Judgement
success All success All success All
total fraction total fraction total fraction
Exp1 (Dataset pair)
Wu-Palmer 772 1660 0.47 464 830 0.56 92 166 0.55
Resnik 859 1660 0.52 514 830 0.62 96 166 0.58
LDA+Wup 674 1700 0.4 506 850 0.6 41 170 0.24
LDA+Res 429 1700 0.25 359 850 0.42 19 170 0.11
BM25 1351 1900 0.71 744 950 0.78 143 190 0.75
Bert_Base 1465 1900 0.77 800 950 0.84 159 190 0.84
LDA 1360 1900 0.72 767 950 0.81 154 190 0.81
Doc2vec 941 1900 0.5 582 950 0.61 144 190 0.76
Exp2 (Query-dataset pair)
Wu-Palmer 83 190 0.44 44 95 0.46 6 19 0.32
Bert_Base 118 190 0.62 66 95 0.69 11 19 0.58
BM25 151 190 0.79 83 95 0.87 17 19 0.89
Resnik 64 190 0.34 39 95 0.41 6 19 0.32
LDA 60 190 0.32 34 95 0.36 8 19 0.42
Doc2vec 123 190 0.65 76 95 0.8 16 19 0.84
Table 3: Scenario 2: Top10, Top5 and Top1 results.(Only consider datasets with subject-area).
Experiment Measure
Top10 Judgement Top5 Judgement Top1 Judgement
success Only-SubjectArea success Only-SubjectArea success Only-SubjectArea
total fraction total fraction total fraction
Exp1 (Dataset pair)
Wu-Palmer 772 1253 0.62 464 760 0.61 92 163 0.56
Resnik 859 1306 0.66 514 795 0.65 96 164 0.59
LDA+Wup 674 1264 0.53 506 839 0.6 41 170 0.24
LDA+Res 429 1126 0.38 359 817 0.44 19 170 0.11
BM25 1351 1715 0.79 744 948 0.78 143 190 0.75
Bert_Base 1465 1736 0.84 800 950 0.84 159 190 0.84
LDA 1360 1694 0.8 767 950 0.81 154 190 0.81
Doc2vec 941 1539 0.61 582 940 0.62 144 190 0.76
Exp2 (Query-dataset pair)
Wu-Palmer 83 155 0.54 44 93 0.47 6 19 0.32
Bert_Base 118 167 0.71 66 95 0.69 11 19 0.58
BM25 151 169 0.89 83 95 0.87 17 19 0.89
Resnik 64 154 0.42 39 95 0.41 6 19 0.32
LDA 60 159 0.38 34 95 0.36 8 19 0.42
Doc2vec 123 150 0.82 76 95 0.8 16 19 0.84
no subject-area in their meta-data. In Table 2, there
is a difference between the "total" number of judge-
ments performed for each approach. This is because
in the concept-based approaches, no concept could be
extracted for some datasets or queries. We have re-
moved these cases from the counts in the table, since
no meaningful similarity score can be computed. Ta-
ble 2 shows that the Bert-based approach outperforms
all others in Exp1, reaching 80%, while LDA-based
approach and BM25-based approaches also perform
well by reaching about 80% accuracy. The table also
shows that the concept-based approaches all seriously
underperform on the task when using datasets as the
original target.
The table also shows the reason why we test they
Hybrid approach by combining LDA with concept-
based approaches. As Table 2 shows, the LDA ap-
proach performs very well on Exp1 (third best perfor-
mance), which means that LDA can extract relatively
correct topics in the majority of cases. This prompted
the hypothesis: extracted topics of datasets by LDA
could be used as concepts to help the concept-based
approach. However, the data in Table 2 shows that this
hypothesis was false: the hybrid approach of LDA
plus either Wu-Palmer or Resnik performs no better
(and sometimes even substantially worse) than Wu-
Palmer or Resnik on their own.
In Exp2 in Table 2, we test 6 approaches with-
out considering the Hybrid approach. Because the
LDA approach doesn’t perform well in Exp2, we do
not expect it to boost the concept-based approaches.
In Exp2, the BM25-based approach outperforms oth-
ers by reaching 85% accuracy. The Bert-based and
Doc2vec-based approaches also perform well. Again,
we see a substantial underperformance of either of the
two concept-based approaches.
Scenario 2. Scenario 2 is a revised version of sce-
nario 1, in which we only consider TopN datasets that
mention the subject-area in their meta-data, so that
we can be certain about the correctness of the rec-
ommendation. For Exp1 of this scenario, the results
of Table 3 show that (again) both the Bert-based and
the LDA-based approach perform well, reaching 80%
accuracy, with the Bert-based approach outperform-
ing others. Also, the BM25-based approach performs
well by reaching 75%. For Exp2, the BM25-based ap-
proach even outperforms all others by reaching about
89% accuracy. The Doc2vec-based approach also
performs well with reaching about 80% accuracy.
Overall, we can conclude as the final result that
BM25-based approach performs best in both Exp1
and Exp2 among all the approaches we tested.
DATA 2021 - 10th International Conference on Data Science, Technology and Applications
198

6 CONCLUSION
In this paper, we provided a novel task for recom-
mending scientific datasets. This task recommends
similar dataset based on a target that is either a query
or another dataset. Based on this task, we intro-
duced several approaches, using some popular sim-
ilarity methods. Also, we executed experiments to
evaluate these approaches on biomedical datasets.
There are a number of lessons that we can draw
from our experiments: 1) it is notable that the task
of recommending similar datasets based on only the
meta-data from these datasets, and possibly a query,
is much harder than one might expect, with even the
best performing methods rarely scoring higher than
80%. 2) we see that the semantic, ontology-based
methods are not capable of solving this task, and that
the statistical methods from machine learning or in-
formation retrieval far outperform the semantic meth-
ods. Even boosting the ontology-based methods with
a machine learning method did not give an accept-
able result. 3) our results show that the BM25-based
approach performs well on the task of dataset recom-
mendation from both a query target and a dataset tar-
get, reaching 70% accuracy in our experiments.
In this paper, we have used only textual meta-data
for dataset recommendation. Given the high vari-
ability in the syntax and semantics of the content of
datasets (ranging from gene sequences to geograph-
ical maps to spreadsheets with financial data), it is
near impossible to use this dataset contents. Never-
theless, there are other signals that could be consid-
ered for similar dataset recommendation. Authors of
datasets would be one of these signals: the co-author
network could be used for dataset recommendation
if we could match dataset’s author into this network.
The Open Academic Graph (OAG) is a very popular
and large knowledge graph that unifies two separate
billion-scale academic graphs MAG and AMiner (Mi-
crosoft, 2021; Sinha et al., 2015; Tang et al., 2008).
We will investigate in future work if this resource can
be exploited to improve the task of recommending
similar datasets that we defined in this paper.
ACKNOWLEDGEMENT
This work has been funded by the Netherlands Sci-
ence Foundation NWO grant nr. 652.001.002 which
is also partially funded by Elsevier. The first author
is funded by by the China Scholarship Council (CSC)
under grant number 201807730060.
REFERENCES
Blei, D. M., Ng, A. Y., and Jordan, M. I. (2003). Latent
dirichlet allocation. Journal of machine Learning re-
search, 3(Jan):993–1022.
Chapman, A., Simperl, E., Koesten, L., Konstantinidis, G.,
Ibáñez, L.-D., Kacprzak, E., and Groth, P. (2020).
Dataset search: a survey. The VLDB Journal,
29(1):251–272.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). Bert: Pre-training of deep bidirectional trans-
formers for language understanding.
Ellefi, M. B., Bellahsene, Z., Dietze, S., and Todorov, K.
(2016). Dataset recommendation for data linking: An
intensional approach. In ESWC 2016, pages 36–51.
Springer.
Google Developers (2021). Dataset | google search
central. https://developers.google.com/search/docs/
data-types/dataset.
Kunze, S. R. and Auer, S. (2013). Dataset retrieval. In 7th
IEEE Int. Conf. on Semantic Computing, ICSC ’13,
page 1–8.
Le, Q. V. and Mikolov, T. (2014). Distributed representa-
tions of sentences and documents. In ICML.
Leme, L. A. P. P., Lopes, G. R., Nunes, B. P., Casanova,
M. A., and Dietze, S. (2013). Identifying candidate
datasets for data interlinking. In Web Engineering,
pages 354–366. Springer.
Microsoft (2021). Open academic graph - microsoft re-
search. https://www.microsoft.com/en-us/research/
project/open-academic-graph/.
Patra, B. G., Roberts, K., and Wu, H. (2020). A
content-based dataset recommendation system for re-
searchers—a case study on Gene Expression Omnibus
(GEO) repository. Database, 2020.
Resnik, P. (1995). Using information content to evaluate se-
mantic similarity in a taxonomy. In IJCAI, IJCAI’95,
page 448–453.
Robertson, S., Walker, S., Jones, S., Hancock-Beaulieu,
M. M., and Gatford, M. (1995). Okapi at trec-3.
In Overview of the Third Text REtrieval Conference
(TREC-3), pages 109–126.
Singhal, A., Kasturi, R., Sivakumar, V., and Srivastava, J.
(2013). Leveraging web intelligence for finding inter-
esting research datasets. In WI-IAT 2013, pages 321–
328. IEEE.
Sinha, A., Shen, Z., Song, Y., Ma, H., Eide, D., Hsu, B.-
J. P., and Wang, K. (2015). An overview of microsoft
academic service (mas) and applications. In WWW
Conference, page 243–246. ACM.
Tang, J., Zhang, J., Yao, L., Li, J., Zhang, L., and Su, Z.
(2008). Arnetminer: Extraction and mining of aca-
demic social networks. In SIGKDD, page 990–998.
ACM.
Wang, X., Huang, Z., and van Harmelen, F. (2020). Evalu-
ating similarity measures for dataset search. In WISE
2020, pages 38–51. Springer.
Wu, Z. and Palmer, M. (1994). Verb semantics and lexical
selection. In 32nd Annual Meeting of the Association
for Computational Linguistics, pages 133–138.
Biomedical Dataset Recommendation
199
