
De-identification of Medical Information for Forming Multimodal
Datasets to Train Neural Networks
Margarita Suzdaltseva
a
, Alexandra Shamakhova
b
, Natalia Dobrenko
c
, Olga Alekseeva
d
,
Jaafar Hammoud
e
, Natalia Gusarova
f
, Aleksandra Vatian
g
and Anatoly Shalyto
h
ITMO University, 49 Kronverksky av., St. Petersburg, Russia
Keywords: Medical Information, De-identification, Multimodal Datasets, Named Entity Recognition, Electronic
Healthcare Record, Rule-based Approach.
Abstract: An important source of medical information for forming multimodal datasets to train neural networks is
electronic patient records. In order to process data from electronic health records with a specified purpose, the
number of requirements must be met - first of all, de-identification. This paper discusses the first stage of this
process - searching for named entities in medical texts (which should be replaced or encrypted afterwards). The
problem is solved by an example of semi-structured EHRs in Russian as a fusional, grammatically complex
language. The structure and specificity of EMC typical for Russia is analyzed in detail. A problem-oriented
comparison of approaches to solving the NER problem is carried out. We developed a pipeline for processing of
HER and experimentally showed the advantages of the rule-based method over using specialized libraries. The
achieved Recall and Precision values were 0.990 and 0.980 respectively.
1 INTRODUCTION
In the modern world, when fighting against new
dangerous diseases takes central stage, development
of medicine would be impossible without advanced
technologies. For instance, application of machine
learning algorithms for clinical data analysis demands
complex datasets containing multimodal information
(including anamnesis, medical images, etc.).
Nowadays necessity of such datasets is perceived not
only at the level of individual universities and funds
who develop large datasets (Armato et al., 2011) but
also at the governmental level (EGISZ, 2017).
Current deficit of high-quality medical datasets is
obvious, since there is a lack of annotated, structured
and pre-processed data. At the same time, a large
amount of unstructured data remains unused. In order
to process data from electronic health records (EHR)
a
https://orcid.org/0000-0002-8136-7925
b
https://orcid.org/0000-0003-4414-4607
c
https://orcid.org/0000-0001-6206-8033
d
https://orcid.org/0000-0001-5976-3393
e
https://orcid.org/0000-0002-2033-0838
f
https://orcid.org/0000-0003-4414-4607
g
https://orcid.org/0000-0002-5483-716Х
h
https://orcid.org/0000-0002-2723-2077
with a specified purpose, the number of requirements
must be met - in particular, de-identification. In other
words, need for EHR data de-identification occurs
when there is a need for any reuse. This is especially
important when creating free datasets on their basis,
which are the basis for the progress of high-tech
medicine.
2 BACKGROUND AND RELATED
WORKS
Legislation of each country interprets the term of de-
identification differently.
In the USA the law in force is the Health
Insurance Portability and Accountability Act 1996.
Here Protected Health Information (PHI) includes
Suzdaltseva, M., Shamakhova, A., Dobrenko, N., Alekseeva, O., Hammoud, J., Gusarova, N., Vatian, A. and Shalyto, A.
De-identification of Medical Information for Forming Multimodal Datasets to Train Neural Networks.
DOI: 10.5220/0010406000002931
In Proceedings of the 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2021), pages 163-170
ISBN: 978-989-758-506-7; ISSN: 2184-4984
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
163

health records, health histories, lab test results, and
medical bills and has 18 identifiers, which should be
de-identified. (Alder, 2017). In the European Union
the law in force is the Regulation (EU) 2018/1725 of
the European Parliament and of the Council 2018. In
the Russian Federation, there are two main laws that
should be relied upon when processing text Federal
Law on the Fundamentals of Protection of the Public
Health 2020 and Federal Law on Personal Data 2006
(edition from 24.04.2020).
It is important to mention that all laws describe
not only a list of information which is considered
personal data, but also requirements for their
processing, storage, transfer, etc. Cryptography must
be used as well, but this is not the focus of the paper.
This paper mostly discusses the stage of searching for
named entities in medical texts (which should be
replaced or encrypted afterwards).
The most relevant and at the same time legitimate
(verified) data on patients' diseases are accumulated
in the Electronic Health Record (EHR), which are the
most valuable source of information in the formation
of multimodal datasets for training neural networks
(Fifty-eighth, 2005).
Naturally, with the advent of EHR, standards were
also formed (Official, June 2019) – for instance,
openEHR (openEHR, 2020), HL7 (HL7
International, 2016) or ASTM Committee E31
(ASTM International, 2020). In addition to the
general structures of the EHR, there are also standards
for medical terms, for example, ICD, approved by
WHO (World Health Organization, 2020). Russia, as
a member of WHO, claims the adherence to these
standards. However, as practice of Russian healthcare
system and our research show, standards are not met
often. In addition, Russian, as a fusional language, is
characterized by complex grammatical constructions,
flexibility, and our EHR characterized by entities,
which are embedded within another entities. All these
factors make solution of NER more complicated.
Named Entity Recognition (NER) approaches are
widely used in processing of medical information.
Researchers outline the following classification of
traditional approaches to NER (Li et al., 2020): rule-
based approaches, unsupervised learning approaches
and feature-based supervised learning approaches.
Each group is characterized by its specific features,
advantages and disadvantages.
Hand-crafted rules are the basis of rule-based
NER systems, especially for EHR (Quimbaya et al.,
2016). The approach is proved to be useful for
improving recall while having limited impact on
precision, which is especially important for the
purposes of this work (see section 3.2).
The basic approach of unsupervised learning is
clustering. Context similarity is the base of named
entities extraction from the clustered groups in
clustering-based NER systems (Alsudais et al., 2018),
which needs the large corpus statistics. Supervised
learning within NER is normally a multi-class
classification or sequence labeling task. Features
should be carefully designed to represent each
training example, considering annotated data samples
(Liu, 2019). Traditionally implemented models are
Hidden Markov Models (HMM), Maximum Entropy
Models (Martino et al., 2018), Support Vector
Machines (SVM) (Gholami et al., 2017), and
Conditional Random Fields (CRF) (Friedrich et al.,
2019).
Nowadays DL-based NER models are becoming
prevalent since they achieve state-of-the-art results
(Hahn et al., 2020). One of the key advantages of deep
learning compared to feature-based approaches is an
ability to discover hidden features and dependencies
automatically (Yadav et al., 2016). Yet there is a main
difficulty in using deep learning for NER (Khin et al.,
2018): to train a supervised NER system is required
to have big annotated data (Lee et al., 2020). In
general, DL-based NER shows poor results on poorly
structured and poorly formatted texts - for example,
on user-generated text e.g., WUT-17 dataset, the best
F-scores are slightly above 40% (Li et al., 2020)
Different approaches are developed to perform
NER on small data. One of them is Active Learning
Query Strategy (Liu et al., 2019), when the machine
learning de-identification system can actively request
information from beyond the system. The best result
shows a Bi-LSTM+CRF architecture in combination
with MultiBPEmb and Flair Multilingual Fast
embeddings, first trained on English data and then on
Italian (Catelli et al., 2020).
Other example of an approach working with a
small amount of data (200 nursing notes) relies on
lookup tables, decision rules and fuzzy string
matching (Menger et al., 2018; Norgeot et al., 2020).
Hybrid approaches also demonstrate good results
(Zhao et al., 2018; Lee et al., 2017).
It is possible to conclude that effective NER
methods for de-identification are already well-known
and widely described in different papers. However,
their choice and implementation in relation to the
realities of Russia analyzed above remain an open
problem.
In order to potentially adapt to a specific task,
authors learned existing software solutions that can be
used to look for named entities in the Russian-
language text.
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
164

One of the most wide-used libraries for Natural
Language Processing (NLP) in Python is SpaCy
(spaCy, n.d). However, there is no adaptation for
Russian language at the moment. Another well-
known library for NLP is Stanza (Stanza, n.d.). In
contradistinction from SpaCy, this library maintains
Russian. An example of a library developed
specifically for Russian language is Natasha
(natasha, n.d.). Natasha solves basic NLP tasks for
Russian language. But none of these libraries are
trained on medical texts, so their direct use on EHR is
problematic.
As reported earlier, security requirements of the
data stored in considered EHR are fairly strict. In
particular, re-identification, i.e. reverse person
identification based on anonymized data, is
unacceptable In this regard, rule-based NER methods
have significant advantages.
On the other hand, the use of Machine Learning
(and Deep Learning, in particular) requires a
significant amount of properly annotated data (Gligic
et al., 2020).
Thus, the analysis showed that in conditions of a
small amount of available medical data, in relation to
the Russian language and the realities of Russian
healthcare, it is advisable to use a rule-based approach
at first, and then try two existing modules for NER in
Russian based on pretrained models instead of
training our own.
Thereby, the aim of this paper is to develop a
pipeline for processing of semi-structured EHRs on
the example of the Russian language as a fusional,
grammatically complex language. It includes the
following subtasks:
1. Preprocessing of a document for further
named entities search,
2. NER implementation,
3. Evaluation of results.
3 METHODS AND MATERIALS
3.1 Characteristics of Gathered Data
EHR provided for the research have characteristics
and properties which are important to discuss.
For the further processing, a dataset was formed
from different types of electronic medical records in
Russian (e.g. medical examinations results,
anamnesis, discharge summary, observation diaries,
etc.) In total they have characteristics presented in
Table 1.
Table 1: Data characteristics.
T
yp
e Characteristic
Words 83 666
Characters 587 264
Formats DOC, PDF, JPG
In general, medical records are not meant for
random people, but for other medical professionals.
This fact explains why language used is very specific:
it includes non-standard and uncommon
abbreviations, short forms, fixed expressions,
medical terms, etc. for example: “KOS” —
“Kislotno-osnovnoe sostoyanie organizma” (The
acid-base state of the body), “LPONP” —
“Lipoproteinov ochen' nizkoj ploskosti” (Very low
plane lipoprotein), “Pron-poziciya” (Prone-position),
“zav. IO” — “zaveduyushchij infekcionnym
otdeleniem” (head of the infectious diseases
department), “lech. vrach” — “lechashchij vrach”
(attending doctor).
Also, the considered records (like almost any text)
are prone to typos — one of the key problems of
working with free-text. Mistakes have been found in
the full name form: “Surname NP” (N - name, P -
patronymic, should be written separately and finish
with dots). Typos may occur in the common lexis, for
example, “nastoshchee” — “nastoyashchee”
(nowadays), where can be corrected with models
trained on any texts, as well as in the functional
medical lexis, for instance, “insuflyaciya” —
“insufflyaciya” (insufflation), where such misprints
can be corrected either manually or with the help of
models, trained especially on large volume of medical
texts, which does not exist for Russian language at the
moment.
The prevalent typo in the provided EHR is the absence
of a space between words:
“Soputstvuyushchij:Gipertonicheskaya” (Concomitant:
Hypertensive), “otdeleniem.Prodolzhaetsya”
(office.Continues) (this typo was made once and
copied several times), “vvide” — v vide (in the form
of). If, in the case of a punctuation mark between
words, it is possible to recognize them as separate
words, then in the case of a missing space between
two words, where the second word is with a small
letter and there are no punctuation marks between,
correction is almost impossible.
Non-standard medical abbreviations (combined
with typos) becomes another problem, so the same
phrase “klinicheskim farmakologom” (clinical
pharmacologist) is written in at least three versions:
“klin.farmakologom”, ”klin.farmakologim”,
”kl.farmakologom”. One more issue with
construction is the lack of dots: “atm dav” —
De-identification of Medical Information for Forming Multimodal Datasets to Train Neural Networks
165
“atmosfernoe davlenie” (atmosphere pressure). There
are also mistakes in dates: in one of the provided
medical records, one patient has three different dates
of birth. All of the above impacts processing. Even
with the right choice of method and writing the code
correctly, the individual entities could not be
extracted.
As a rule, the EHR from the examined sample
have no strict logical structure. It is still possible to
distinguish individual structural elements in it, like
tables of laboratory results, discharge reports, patient
observation diaries, and other notes in electronic
form. Though, blocks are not ordered, alternate
chaotically, do not have hyperlinks to each other.
Connections are visible on close reading, but medical
card structure does not reflect them. For example, a
test was taken on one of the days of hospitalization,
but the conclusions drawn from its results are at the
other end of the electronic medical record.
Furthermore, understanding the structure of
information was hampered by multiple repetitions of
the same data. Sometimes it was not clear if the result
was similar to the previous one or simply duplicated.
As already noted, although integral EHR are being
actively implemented in large cities of Russia,
electronic medical records without clear structure and
even conventional handwritten medical records are
still in use. Widespread standards OpenEHR и HL7
(and other standards mentioned above) are not met
often. This is also true for the given documents. In
fact, an EHR in this case is a document consisting of
hand-printed medical texts (narrative, statement of
facts), tables (or pictures) with test results in
electronic format. Obviously, data has neither
annotation nor appropriate markup.
All the above factors make it difficult to perform
NER. However, it is undeniable that the EHR
described potentially contain useful information to be
used while preparing a dataset (e.g. for predicting
disease outcome). Nevertheless, de-identification is a
necessary step in medical data reuse. Consequently, it
was required to find an optimal way of realization.
3.2 Metrics and Baseline
We used metrics that are traditional for NER
solutions, namely:
##
Precision = ; Recall = ;
#( ) #( )
TP TP
TP FP TP FN
Precision Recall
2
Precision Recall
Fscore
.
All EHR under consideration have been annotated
manually. If surname were obtained in several
grammatical categories, all forms were considered as
different surnames.
It is worth note that in the task of de-identification,
recall is more important, since the main goal is safety
of patient personal data. It is necessary to recognize
all named entities present in the text. A single pass
can lead to re-identification: even one doctor’s
surname left in the text can easily disclose the medical
institution where they work and re-identify patients
(number of patients is probably limited). False
positives are allowed, precision does not have to be
as high. Accordingly, in our task to erase false
positives is better than to leave false negatives
(unrecognized entities).
The risk of re-identification is real and can lead to
serious breaches of patient privacy and
confidentiality. As noted in (Yogarajan et al., 2019),
while designing an automatic de-identification
system, it is important to consider the re-
identification risk and take appropriate measures to
minimize such risk.
As the analysis of the literature shows, many
researchers focus on accuracy (and prioritize raising
accuracy above the standard value of 95%), but it is
also important to analyze de-identified text and take
qualitative factors into consideration. Obviously, in
addition to a quantitative assessment of the results of
de-identification, its qualitative expert assessment
should be carried out, however, it is already expedient
to solve this task after the complex processing of the
EHR text, including the replacement of the identified
TCA, which is beyond the scope of this article.
As a baseline for comparison, we chose a rule-
based system for automatic de-identification of
medical narrative texts for Serbian (Jaćimović et al.,
2015), since it, like Russian, belongs to the group of
Slavic languages and has similar problems.
4 RESULTS AND DISCUSSION
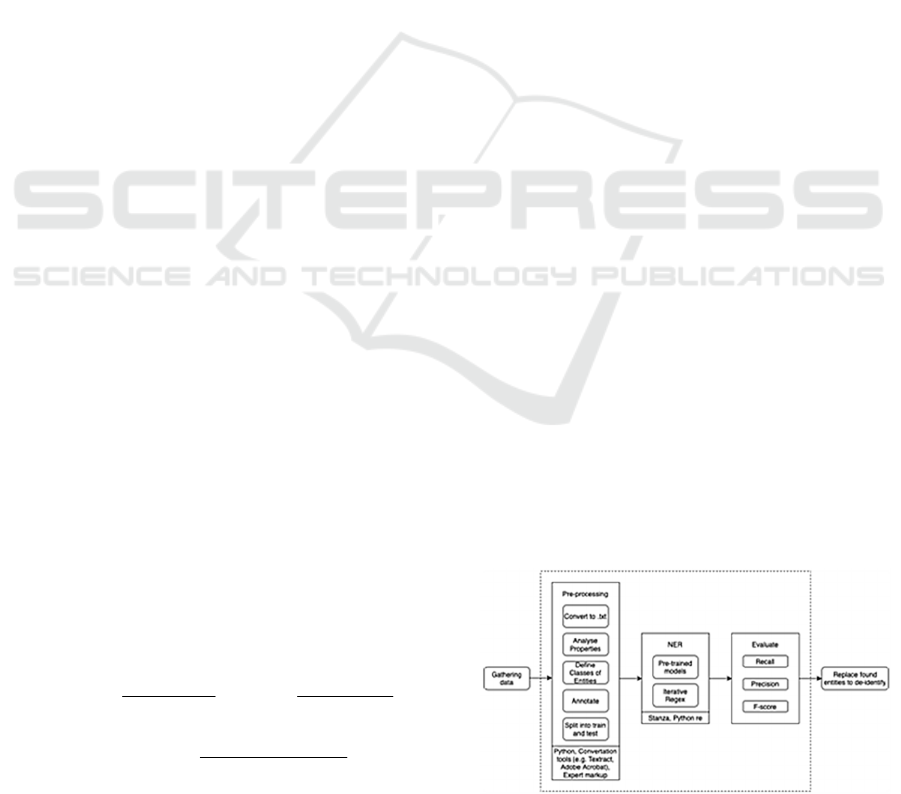
Figure 1: Pipeline of the proposed solution.
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
166

The developed pipeline for processing of EHRs is
presented in Figure 1. Area of responsibility of our
article lies within the dashed rectangle.
4.1 Preprocessing
In order to preprocess data authors analyzed the
structure of the documents. EHRs contain a variety of
multimodal medical information. Raw data was
received in .doc, .pdf, .jpg formats. High-tech images
tend to be in native formats such as DICOM, however
due to the transfer from a third-party program for
EHR maintenance, scans were in .pdf and .jpg
formats.
Original documents did not have precise logical
structure. In .doc files text (also numbers, tables) was
contained in nested tables. To solve problems
associated with hierarchical nesting structure of a
document, .doc files were converted into .txt with
Python package
textract. This expectedly caused
loss of original structure (e.g. footers and headers),
the appearance of blank lines, etc., but all key
information was saved. .pdf can be easily converted
to any other format. .jpg files were converted to .pdf
at first. It can be performed with Adobe Acrobat or
some other application using OCR to recognize text
on an image. Documents in the .txt format were ready
to be subjected to program processing.
Authors of the paper revealed that texts contained
syntax and grammatical errors, mistakes, typos. Their
number is roughly 87 errors per 10 thousand
characters of text.
Table 2: Entities.
Entity class Amount
p
erson
(
uni
q
ue
)
180
or
g
anization
(
uni
q
ue
)
17
id numbers
(
uni
q
ue
)
13
location (unique) 2
date (all) 215
all 427
To be able to evaluate results further, the dataset
was split into train and test in a ratio of 2 to 1. Next,
classes of entities were revealed: person (patient,
doctors, laboratory assistants...), organization
(medical center name, polyclinics), id-numbers
(identity documents; policy number; insurance
number; number of issued certificate of incapacity for
work; EHR number), location (addresses), date
(patient birth date; dates of appointments, observation
diaries, test results). The list of classes was formed in
accordance with Russian Federal Law on the
Fundamentals of Protection of the Public Health
2020. Comparison of entities given in law and entities
which actually contained in EHR allowed to form the
final list of classes (Table 2).
Finally, manual expert markup of the train and test
samples was performed.
4.2 NER Implementation
4.2.1 Iterative Regex
A rule-based approach was chosen as the main one
for de-identification of EHRs in Russian. Authors
used the programming language Python. So, an
iterative approach to search and replacement of
named entities in the text was developed, namely:
necessary regular expressions for each class should
not be used all at once, but in a specific order. At each
iteration we search in the text, that does not store
original forms of entities found at the previous
iteration. This technique may be useful, for example,
if you need to keep the roles of personalities in the
text, which is especially important when forming
complex Multimodal Datasets.
A decision tree reflecting optimal order of entities
(their search and replacement) was built within this
method. The order of iterations authors suggested:
1. Organization & location
2. Id numbers
3. Person (patient)
4. Person (the rest)
5. Date (birthday)
6. Date (the rest)
These regular expressions are used to look for
combinations of first name, last name and patronymic
within rule-based approach. Separate regular
expressions for different forms to record names allow
us to take a position of surname into account and
extract it separately if needed.
The listing of full name extraction with Regex is
given in Appendix.
Dates processing depends on de-identification
purposes. For the purpose of compiling a dataset, the
age can be replaced by the age group (the date of birth
is deleted). If the chronology inside the medical card
is not important to us, the date is replaced with a
quarter of a year. If it is crucial to take into account a
chronology inside one medical card, the date of
patient admission is considered as a starting point for
other dates in the card and they are replaced with a
positive or negative number. If it is decisive to
understand how a batch of health records is
chronologically arranged, then a dictionary of dates is
created, where the start and end dates are customized.
De-identification of Medical Information for Forming Multimodal Datasets to Train Neural Networks
167

Deleting of document numbers, passport data,
insurance, policy, addresses and phone numbers,
email addresses is done with regular expressions,
because all these numbers have templates. For
instance, the passport number consists of ten digits.
Besides, such entities have unique symbols: “№”,
“@”, “:” (Colons also appear in other personal data,
hence it is important to keep the chronology of de-
identification).
The listing of dates parsing is given in Appendix.
4.2.2 Pre-trained Models
The next method considered in this section - to use
appropriate pretrained models for NER. Packages
were used with Python language.
Natasha library has a NER module using pre-
trained neural networks. The entity classes person,
location, organization have been filtered.
Stanza library is also NER based on ML. The
entity classes person, location, organization have
been extracted.
Entities date and id numbers have been extracted
only with rule-based approach (regular expressions),
not NER, since the recording of dates is rather strictly
formalized, and id numbers were encountered in a
limited number of contexts.
The Listings of the extraction named entities with
Natasha library, as well as of the txtraction of entity
classes person, location, organization (‘PER’, ‘LOC’,
‘ORG’) with Stanza library are given in Appendix.
4.3 Evaluation
As a result of applying selected de-identification
approaches on test data we got metrics to compare
with the baseline.
Table 3: Metrics.
method \ metrics Recall Precision F-score
Baseline 0,96 0,97 0,97
Rule-
b
ase
d
0,990 0,980 0,985
Stanza 0,941 0,842 0,888
Natasha 0,714 0,820 0,760
As you can see from the Table 3, the Rule-based
method showed the best results both in comparison
with the baseline and in comparison with the Pre-
trained models. Noteworthy is the high Recall value
obtained by this method, which almost eliminates the
need for additional manual (expert) verification of de-
identified EHRs.
However, each of the proposed solutions is not
free from disadvantages.With an iterative rule-based
approach, problems arise with every new form of
notation or misspellings in letter case or punctuation
marks.
Natasha's problem is that it does not find non-
standard surnames, at the same time it captures the
names of drugs or substances that are important for
the further use of information.
Stanza successfully coped with the definition of
named entities, has high recall and the percentage of
captured false positive values is relatively small. On
the other hand, if the names of locations and
organizations are extracted, the estimates will
deteriorate significantly. False Positive entities could
be removed with regular expressions.
Thus, there are prerequisites for using a hybrid
approach to improve the efficiency of NER.
5 CONCLUSIONS
The paper discussed the problem of de-identification
of medical information for forming multimodal
datasets to train neural networks (on example of EHR
in Russian). The analysis of the legislation of various
countries in relation to the problem under
consideration is carried out, the difficulties of solving
the NEP problem in relation to EHR in Russia are
revealed, both from the point of view of compliance
with the law and from the point of view of the
specifics of the Russian language. A problem-
oriented comparison of approaches to solving the
NER problem was carried out.
The tasks set in the article have been successfully
solved, namely, the pipeline for processing EHR was
developed. Its main stages are preprocessing, NER
implementation, and final evaluation. A fairly high
efficiency of the proposed solution is shown
experimentally: the achieved Recall and Precision
values were 0.990 and 0.980 respectively.
After the processing described, textual data can be
applied to formation of multimodal datasets
appropriate for training neural networks.
ACKNOWLEDGEMENTS
This work was financially supported by Russian
Science Foundation, Grant 19-19-00696, and Grant
of the President of the Russian Federation for state
support of young Russian scientists - candidates of
science, MK-5723.2021.1.6.
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
168

REFERENCES
Armato, S.G., McLennan, G., Bidaut, L. et al. (2011). The
Lung Image Database Consortium (LIDC) and Image
Database Resource Initiative (IDRI): a completed
reference database of lung nodules on CT scans.
Medical Physics, 38(2), 915-31. doi: 10.1118/
1.3528204
EGISZ, Unified state information system in the field of
healthcare 2017 (№ 242-FZ) (Russia)
The Health Insurance Portability and Accountability Act
1996 (HIPAA) (U.S.)
Alder, S. (2017). What is Considered PHI Under HIPAA?
HIPAA Journal. https://www.hipaajournal.com/
considered-phi-hipaa/
Alsudais, A. & Tchalian, H. (2018). Clustering Prominent
Named Entities in Topic-Specific Text Corpora. CoRR.
arXiv:1807.10800
ASTM International. (n.d.). Committee E31 on Healthcare
Informatics. Retrieved on September 2, 2020, from
https://www.astm.org/COMMITTEE/E31.htm
Catelli, R., Gargiulo, F., Casola, V., Pietro, G., Fujita, H. &
Esposito, M. (2020). Crosslingual named entity
recognition for clinical de-identification applied to a
COVID-19 Italian data set. Applied Soft Computing,
97(A), ISSN 1568-4946. doi:10.1016/j.asoc.2020.
106779
Federal Law on Personal Data 2006 (№ 152-FZ) (Russia)
Federal Law on the Fundamentals of Protection of the
Public Health 2020 (№ 303-FZ) (Russia)
Fifty-eighth World Health Assembly 2005 (WHA58.28)
https://apps.who.int/iris/bitstream/handle/10665/20378
/WHA58_28-en.pdf?sequence=1
Friedrich, M., Köhn, A., Wiedemann, G., & Biemann, C.
(2020). Adversarial learning of privacy-preserving text
representations for de-identification of medical records.
Paper presented at the ACL 2019 - 57th Annual Meeting
of the Association for Computational Linguistics,
Proceedings of the Conference, 5829-5839.
Gholami, R. & Fakhari, N. (2017). Chapter 27 - Support
Vector Machine: Principles, Parameters, and
Applications. Handbook of Neural Computation, 515-
535, ISBN 9780128113189. doi.org:10.1016/B978-0-
12-811318-9.00027-2
Gligic, L., Kormilitzin, A., Goldberg, P., & Nevado-
Holgado, A. (2020). Named entity recognition in
electronic health records using transfer learning
bootstrapped neural networks. Neural Networks, 121,
132-139. doi:10.1016/j.neunet.2019.08.032
Guidance on De-identification of Protected Health
Information 2012 (U.S.) https://www.hhs.gov/hipaa/
for-professionals/privacy/special-topics/de-
identification/index.html
Hahn, U., & Oleynik, M. (2020). Medical information
extraction in the age of deep learning. Yearbook of
Medical Informatics, 29(1), 208-220. doi:10.1055/s-
0040-1702001
Health Insurance Portability and Accountability Act 1996
(HIPAA) (U.S.)
HL7 International. (2016, May 11). Electronic Health
Records. https://www.hl7.org/Special/committees/ehr/
overview.cfm
Jaćimović, J., Krstev, C., & Jelovac, D. (2015). A rule-
based system for automatic de-identification of medical
narrative texts. Informatica (Slovenia), 39(1)
, 45-53
Khin, K., Burckhardt, P. & Padman, R. (2018). A Deep
Learning Architecture for De-identification of Patient
Notes: Implementation and Evaluation. The 28th
Workshop on Information Technologies and Systems.
arXiv:1810.01570v1
Lee, HJ., Wu, Y., Zhang, Y., Xu, J., Xu, H. & Roberts, K.
(2017). A hybrid approach to automatic de-
identification of psychiatric notes. Journal of
Biomedical Informatics, 75, S19-S27, ISSN 1532-0464,
doi.org:10.1016/j.jbi.2017.06.006
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., &
Kang, J. (2020). BioBERT: A pre-trained biomedical
language representation model for biomedical text
mining. Bioinformatics, 36(4), 1234-1240.
doi:10.1093/bioinformatics/btz682
Li, J., Sun, A., Han, J. & Li, C. (2020). A Survey on Deep
Learning for Named Entity Recognition. IEEE
Transactions on Knowledge and Data Engineering.
arXiv:1812.09449v3
Li, M., Scaiano, M., El Emam, K., & Malin, B. A. (2019).
Efficient Active Learning for Electronic Medical
Record De-identification. AMIA Joint Summits on
Translational Science proceedings. AMIA Joint
Summits on Translational Science, 2019, 462–471.
Liu, S., Sun, Y., Li, B., Wang, W. & Zhao X. (2019).
HAMNER: Headword Amplified Multi-span Distantly
Supervised Method for Domain Specific Named Entity
Recognition. Biomedical NER and Relation
Construction. arXiv:1912.01731v1
Martino, A. & Matrion, D. (2018). An introduction to the
maximum entropy approach and its application to
inference problems in biology. Heliyon, 4(4).
doi.org:10.1016/j.heliyon.2018.e00596
Menger, V., Scheepers, F., Wijk, L. M. & Spruit, M. (2018).
DEDUCE: A pattern matching method for automatic
de-identification of Dutch medical text. Telematics and
Informatics, 35(4), 727-736, ISSN 0736-5853,
doi.org:10.1016/j.tele.2017.08.002
Norgeot, B., Muenzen, K., Peterson, T.A. et al. (2020).
Protected Health Information filter (Philter): accurately
and securely de-identifying free-text clinical notes. npj
Digit. Med. 3, 57. doi.org:10.1038/s41746-020-0258-y
Official Website of The Office of the National Coordinator
for Health Information Technology (ONC). (2019,
September 10). What is an electronic health record
(EHR)? https://www.healthit.gov/faq/what-electronic-
health-record-ehr
Official Website of The Office of the National Coordinator
for Health Information Technology (ONC). (2019, June
4). Health IT Standards. https://www.healthit.gov/
topic/standards-technology/health-it-standards
openEHR. (n.d.). What is openEHR? Retrieved on
September 2, 2020, from https://www.openehr.org/
about/what_is_openehr
De-identification of Medical Information for Forming Multimodal Datasets to Train Neural Networks
169

Quimbaya, A. P., Múnera, A. S., Rivera, R. A. G.,
Rodríguez, J. C. D., Velandia, O. M. M., Peña, A. A. G.
& Labbé, C. (2016). Named entity recognition over
electronic health records through a combined
dictionary-based approach. Procedia Computer
Science, 100, 55–61.
Regulation (EU) 2018/1725 of the European Parliament
and of the Council of 23 October 2018 on the protection
of natural persons with regard to the processing of
personal data by the Union institutions, bodies, offices
and agencies and on the free movement of such data,
and repealing Regulation (EC) No 45/2001 and
Decision No 1247/2002/EC (EU)
spaCy. (n.d.). Industrial-Strength Natural Language
Processing in Python. Retrieved on September 12,
2020, from https://spacy.io
Stanza. (n.d.). Stanza – A Python NLP Package for Many
Human Languages. Retrieved on September 15, 2020,
from https://stanfordnlp.github.io/stanza/
World Health Organization. (n.d.). International Statistical
Classification of Diseases and Related Health
Problems (ICD). Retrieved on September 2, 2020, from
https://www.who.int/standards/classifications/classific
ation-of-diseases
Yadav, S., Ekbal, A., Saha, S., & Bhattacharyya, P. (2016).
Deep Learning Architecture for Patient Data De-
identification in Clinical Records.
ClinicalNLP@COLING 2016.
Yogarajan, V., Pfahringer, B. & Mayo, M. (2019).
Automatic end-to-end De-identification: Is high
accuracy the only metric? Applied Artificial
Intelligence. arXiv:1901.10583v1
Zhao, YS., Zhang, KL., Ma, HC. & Li, K. (2018).
Leveraging text skeleton for de-identification of
electronic medical records. BMC Med Inform Decis
Mak 18, 18. doi.org:10.1186/s12911-018-0598-6
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
170
