
Graph Database on Medical Research Data for Integrated Life Science
Research
Aly Lamuri
1
, Randy Sarayar
2
, Jonathan Aditama Midlando Purba
3
and Adrian Reynaldo Sudirman
2
1
School of Computing, Newcastle University, Newcastle upon Tyne, U.K.
2
Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
3
Menteng Sub-District Primary Healthcare, Jakarta, Indonesia
Keywords: Computing Methodology, Database Management System, Medical Informatics.
Abstract: Indonesia has experienced an increasing surge of published scientific articles in recent years. In medical
science, published articles greatly vary from both pre-clinical and clinical studies, where each study possesses
a different methodological approach and hypothetical premise. However, some articles do not include
rigorous documentation to make it reproducible. Moreover, the lack of centralized database further impedes
researcher from reanalysing previous findings and integrating them with the new study. This paper delineates
such an issue by constructing a graph database to centralize and integrate clinical research data. The database
is constructed using Neo4j and cypher querying language and populated with 5,000 medical records generated
by the synthea program. We address the viabilities of our proposed data curation method by simulating data
of different sizes. Our database was able to answer queries requiring complex relationships while minimizing
the amount of database hits. We conclude that graph databases are quite performant for solving data
integration and centralization issues faced by life science research institutes.
1 INTRODUCTION
Scientific publications in Indonesia have undergone
manifold increases within the past decades. As
reported by Maula, Fuad and Utarini (2018), numbers
of published articles on dengue-related subjects
increased 13 times in 2017, as compared to 2007.
Such an increase was also followed by h-index
improvement, resulting in Indonesia being placed as
the 5
th
most scientifically productive ASEAN country
in investigating dengue-related topics (Maula, Fuad,
and Utarini, 2018). Another bibliometric analysis
investigated by Sarwar and Hassan (2015) also
enlisted Indonesia within 11 of the most scientifically
productive Islamic countries. However, these articles
often did not provide a robustly elaboration of the
methodological procedure or provide the obtained
data for reanalysis; these two factors contribute to
reproducibility and credibility in scientific
publication (Pashler and Wagenmakers, 2012; Stark,
2018; Resnik and Shamoo, 2016). Besides enabling
preprint access (Oakden-Rayner, Beam and Palmer,
2018) and thorough documentation on methodology,
data availability is also a crucial component for
reproducibility in science (Peng, 2015). Therefore,
we proposed utilizing graph databases to integrate
research findings in life science-related fields.
1.1 Graph Database
Data management systems should appropriately
consider interoperability and scalability, which
enable data storing, indexing and retrieving.
Databases aggregate integrated objects in a structure
defined by its metadata. The presence of metadata
implies a self-defined property of the database,
whereas in a relational database management system
(RDBMS), such a definition is included within its
particular schema (Berg, Seymour and Goel, 2012).
During the development of RDBMS, the need to
quickly retrieve the data through a syntactically and
logically feasible manner, therefore inducing the
conceptual design of SQL, a structured querying
language is emerging.
However, with data being stored in a multi-tabular
layout, the relational database (RDB) faced massive
disadvantages in handling highly-connected data.
Hence the development of a schema-less database
initiated by NoSQL (Berg, Seymour, and Goel, 2012;
Fabregat et al., 2018), with a graph database being
Lamuri, A., Sarayar, R., Purba, J. and Sudirman, A.
Graph Database on Medical Research Data for Integrated Life Science Research.
DOI: 10.5220/0009387000050011
In Proceedings of the 4th Annual International Conference and Exhibition on Indonesian Medical Education and Research Institute (The 4th ICE on IMERI 2019), pages 5-11
ISBN: 978-989-758-433-6
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
5
one of its variants (Oussous et al., 2015).
A graph database is more performant in storing
data with intricate relationships (e.g.., protein
interactions or chemical reaction pathways), as
compared to its RDB counterparts (Fabregat et al.,
2018). Neo4j is a graph database platform developed
in Java and compliant towards an ACID system
(Atomicity, Consistency, Isolation, Durability)
(Oussous et al., 2015). As a native graph database,
Neo4j stores data as explicitly defined relationships
in a schema-less management system. Therefore,
Neo4j treats database querying as a graph traversing
process. This redeeming feature of the graph
database, in general, enables higher performance and
flexibility in storing the data. Neo4j employs cypher
as a querying language to define patterns on
traversing the relationship graph. Furthermore, the
ASCII-Art syntax of cypher enables a more intuitive
querying process. Such uniquely written language
and ACID-compliant platforms could become a two-
fold advantages to use Neo4j in delivering a graph
database management system.
1.2 Medical Informatics
Information in the life science-related fields often
possesses an intelligible relationship of a causative
nature. Such information may present a connection
between one entity to another. Interractome,
reactome and connectome are common examples we
see (Figure 3) that may be found in currently
emerging basic science research. In the translational
research paradigm, some interests highlighted the
importance of genetic and proteomic interaction
networks. Meanwhile, in clinical settings, we may
also want to consider patient, doctor and institution as
separate-yet-related entities. Therefore, the nature of
the data in medicine actually closely resembles
entity-relationship data. We will undoubtedly
consider applying a graph database as an alternative
to RDB for use in storing life science-related research
data.
2 METHOD
This study utilized a machine with Intel Core i7-
7700HQ, 8GB of DDR4 RAM and a 5400 RPM
spinning hard disk. We employed Neo4j as a platform
to create a graph database with Cypher as the
querying language. Data used in this study are
generated from a synthea program, producing 5,000
to 50,000 medical records in json-based FHIR (Fast
Healthcare Interoperability Resources), whichwas
directly converted into *.csv format. As shown in
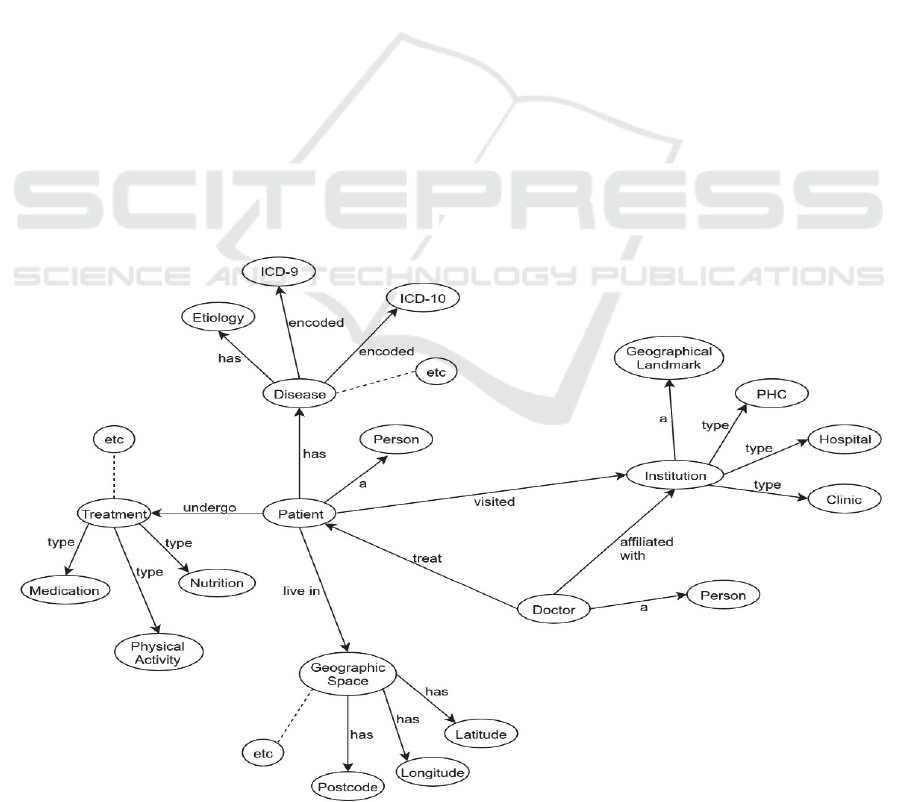
Figure 1, we treated each entity as a vertex and
underlying relationship as an edge connecting two
Figure 1: Schematic representation on graph database for medical records.
The 4th ICE on IMERI 2019 - The annual International Conference and Exhibition on Indonesian Medical Education and Research Institute
6
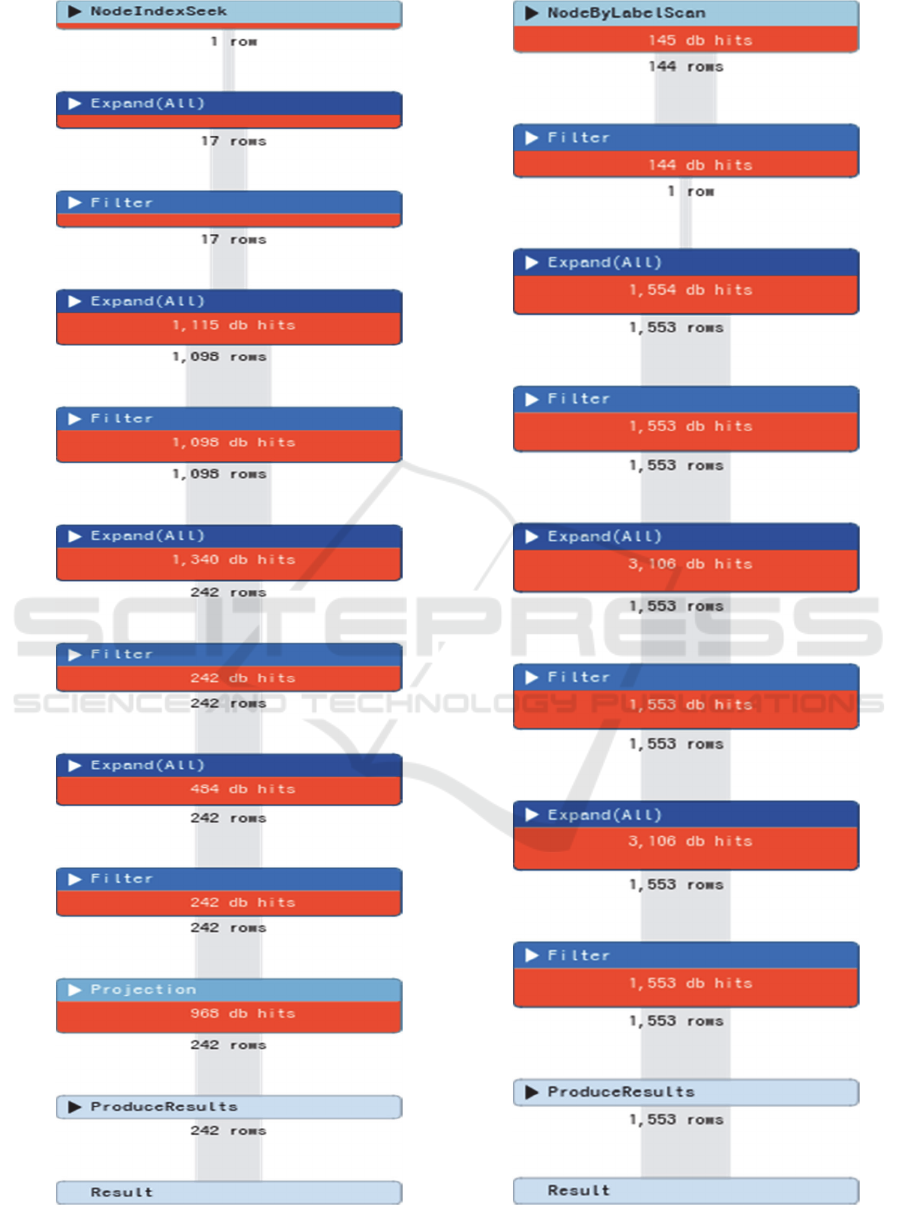
Figure 2: Database hits on the first query.
Figure 3: Database hits on the second query.
Graph Database on Medical Research Data for Integrated Life Science Research
7
vertices. We first designed constraints for unique
input and indices for redundant vertices. To prevent
random access memory (RAM) bottleneck, we
enabled periodic commit for each of the 500 inputs,
which was especially beneficial when dealing with
numerous entries. Afterwards, we loaded a *.csv file
generated by synthea as a query object and set the
entity and relationship.
To measure the performance of our proposed
database, we created a log containing time
consumption and a number of created objects, which
included nodes, relationships, graph property and
graph labels. Said database model took data of
various sizes as input: 5,000, 10,000, 20,000 and
50,000. Considering exponential increment in our
data, we applied power transformation according to
the Tukey ladder of power to normalize the data. The
Anderson-Darling test was then employed to
challenge normality assumption. We computed
correlation estimates between time and created
objects based on p-value obtained from a normality
test. Kendall’s tau estimates the correlation when any
of imputed variable has p < 0.05; Pearson’s is used in
other cases. Data was fitted into a generalized linear
model (GLM) with the Gaussian link function. The
simulation process involved two different queries on
all datasets.
Figure 4: Queries on different datasets. Top: Data size and elapsed time. Bottom: Database hits and elapsed time.
The 4th ICE on IMERI 2019 - The annual International Conference and Exhibition on Indonesian Medical Education and Research Institute
8
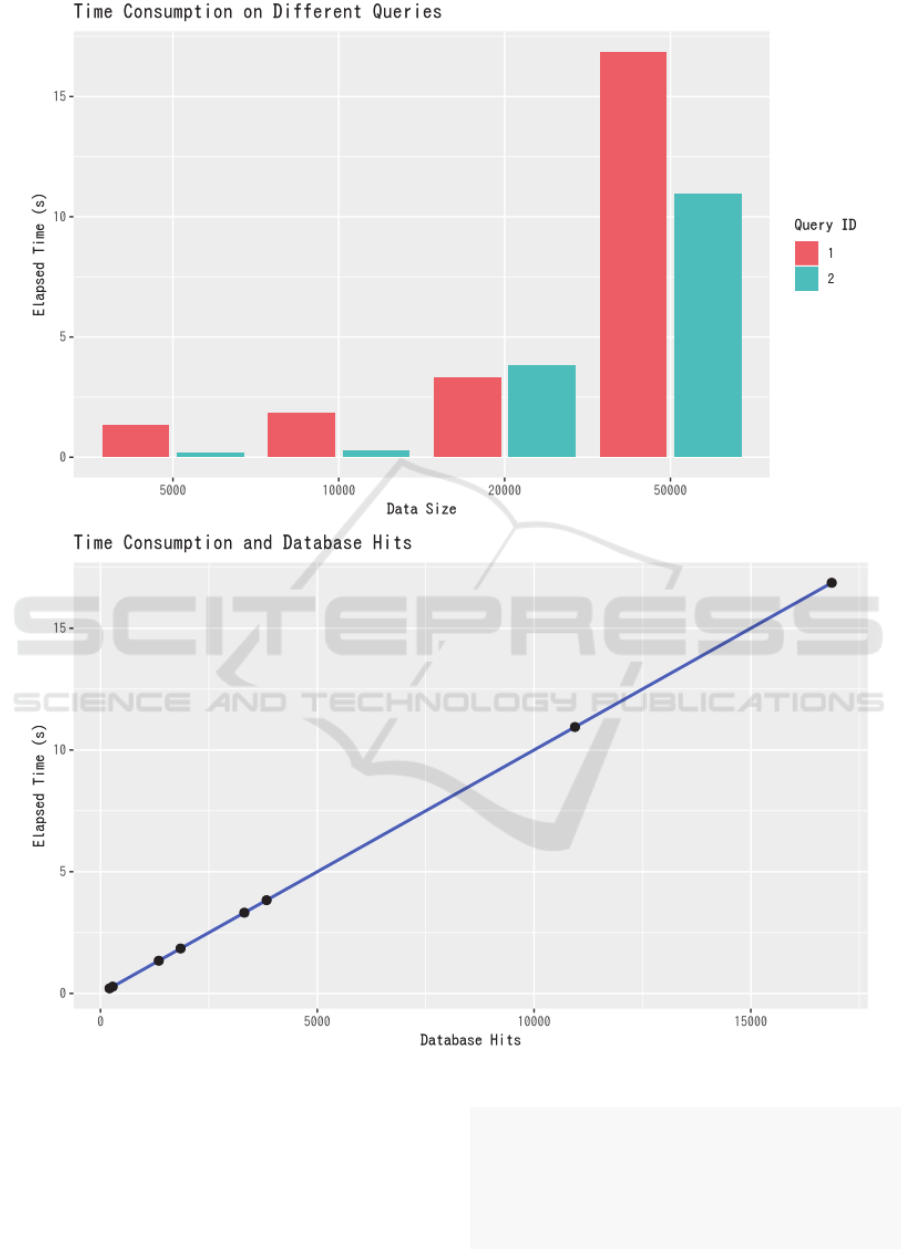
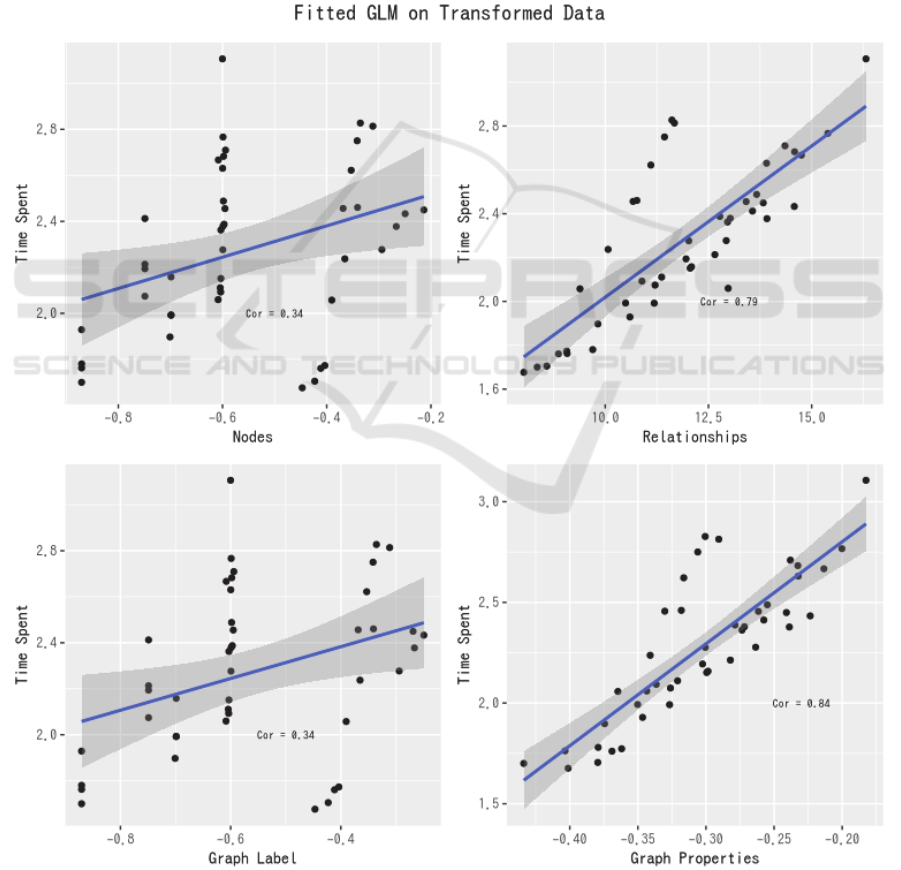
Figure 5: Data transformation and correlation.
Database hits (db-hits) and time measured the
efficacy in handling such queries. Results on
simulation presented in bar plot to demonstrate
database scalability. Queries are written in cypher and
are presented as follows:
// List diagnoses in Massachusetts
match (p:Patient) -[:ATTENDED_AN]->
(e:Encounter) <-[:PROVIDED_AN]-
(o:Organization) -[:LOCATED_IN]-> (g
:GeoLoc)
match (d:Diagnoses) <-[r:HAS_DIAGNOSES]
- (e:Encounter)
Graph Database on Medical Research Data for Integrated Life Science Research
9

return p.Name as Patient,
d.Name as Diagnoses,
o.Name as Institution,
g.Name as City,
r.Date as Date
;
// List patients with hypertension
match (p:Patient) -[:ATTENDED_AN]-> (e:
Encounter)
match (:Diagnoses {Name:'Hypertension'}
) <-[:HAS_DIAGNOSES]-
(e) <-[:PROVIDED_AN]- (o:Organizati
on)
return p, e, o
;
3 RESULT
The constructed database was able to return answers
to queries requiring complex relationships. Our
previous queries assume data with complex
relationships, where each returned a network of
patient, institution and the encounter. Figure and
depicted profile of database hit from both queries.
Depicted in the figure is the representation of query
scalability on data with different sizes. Fitted GLM is
presented as Figure 4, which shows that relationship
and graph property have the most implication on data
input runtime (ρ = 0.79, ρ = 0.84).
4 DISCUSSION
Our study demonstrates the graph database as a
potential platform to store life science research data.
Previous studies emphasized graph database
credibility in storing interconnected data, where a
graph database pattern query on such data may
outperform RDB (Medhi and Baruah, 2017; Fabregat
et al., 2018; Mathew and Kumar, 2014). However, in
other cases requiring analytical query, RDB
outperformed the graph database; in their study,
Hölsch, Schmidt and Grossniklaus (2017) argued that
Neo4j became less performant due to a less advanced
disk and buffer management, compared to RDB. We
therefore conclude that a graph database is quite
performant for integrating medical health records
generated for 5,000 subjects using a synthea program.
Our simulation demonstrated the viability of
storing and querying a large dataset. On exponentially
increasing data size, time consumption on particular
queries also increased exponentially, as demonstrated
in Figure 5. However, it appears to us that further
optimization should be of essence, considering that
query runtime increases from 20,000 to 50,000
dataset. In preparing the database, the log captured
objects, thus causing an immense burden during data
input. Said objects include relationship and graph
property, where previously mentioned graph database
stores an object explicitly instead of implying the
relationship. This feature aids graph database to
answer queries for complex relationships. As such,
longer time spent in creating an object within the
database will not be an issue. During data preparation,
we observed a longer time duration in bigger dataset.
It seems Neo4j may perform better when using
smaller data, so we suggest dividing data into smaller
chunks to improve data input performance.
5 CONCLUSIONS
As a concluding remark, the graph database is quite
performant to integrate medical health record
generated for 5,000 to 50,000 subjects using synthea.
REFERENCES
Berg, K. L., Seymour, T. L., and Goel, R. L. (2012). History
of Databases. International Journal of Management &
Information Systems (IJMIS), 17(1), 29–36.
http://doi.org/10.19030/ijmis.v17i1.7587.
Fabregat, A., Korninger, F., Viteri, G., Sidiropoulos, K.,
Marin-Garcia, P., Ping, P., … Hermjakob, H. (2018).
Reactome graph database: Efficient access to complex
pathway data. PLoS Computational Biology, 14(1),
e1005968. http://doi.org/10.1371/journal.pcbi.1005968.
Hölsch, J., Schmidt, T., and Grossniklaus, M. (2017). On
the Performance of Analytical and Pattern Matching
Graph Queries in Neo4j and a Relational Database. In
Workshop Proceedings of the EDBT/ICDT 2017 Joint
Conference. Venice.
Mathew, A. B., and Madhu Kumar, S. D. (2014). An
Efficient Index Based Query Handling Model for
Neo4j. International Journal of Advances in Computer
Science and Technology (IJACST), 3(2), 12–18.
Maula, A. W., Fuad, A., and Utarini, A. (2018). Ten-years
trend of dengue research in Indonesia and South-east
Asian countries: a bibliometric analysis. Global Health
Action, 11(1), 1504398. http://doi.org/10.1080/
16549716.2018.1504398.
Medhi, S., and Baruah, H. K. (2017). Relational database
and graph database: A comparative analysis. (JPMNT)
Journal of Process Management – New Technologies,
5(2), 1–9. http://doi.org/10.5937/jouproman5-13553.
Oakden-Rayner, L., Beam, A. L., and Palmer, L. J. (2018).
Medical journals should embrace preprints to address
the reproducibility crisis. International Journal of
Epidemiology, 47(5), 1363–1365. http://doi.org/
10.1093/ije/dyy105.
The 4th ICE on IMERI 2019 - The annual International Conference and Exhibition on Indonesian Medical Education and Research Institute
10

Oussous, A., Benjelloun, F.-Z., Lahcen, A. A., and Belfkih,
S. (2015). Comparison and Classification of NoSQL
Databases for Big Data. In Proceedings of International
Conference on Big Data, Cloud and Applications.
Tetuan, Morocco.
Pashler, H., and Wagenmakers, E.-J. (2012). Editors’
Introduction to the Special Section on Replicability in
Psychological Science: A Crisis of Confidence?
Perspectives on Psychological Science, 7(6), 528–530.
http://doi.org/10.1177/1745691612465253.
Peng, R. (2015). The reproducibility crisis in science: A
statistical counterattack. Significance, 12(3), 30–32.
http://doi.org/10.1111/j.1740-9713.2015.00827.x
Resnik, D. B., and Shamoo, A. E. (2017). Reproducibility
and Research Integrity. Accountability in Research,
24(2), 116–123. http://doi.org/10.1080/08989621.
2016.1257387.
Sarwar, R., and Hassan, S.-U. (2015). A bibliometric
assessment of scientific productivity and international
collaboration of the Islamic World in science and
technology (S&T) areas. Scientometrics, 105(2), 1059–
1077. http://doi.org/10.1007/s11192-015-1718-z.
Stark, P. B. (2018). Before reproducibility must come
preproducibility. Nature, 557(7707), 613.
http://doi.org/10.1038/d41586-018-05256-0.
Graph Database on Medical Research Data for Integrated Life Science Research
11
