
Understanding of Non-linear Parametric Regression and Classification
Models: A Taylor Series based Approach
Thomas Bocklitz
1,2
1
IPC Junior Research Group ’Statistical Modelling and Image Analysis’,
Institute of Physical Chemistry and Abbe Center of Photonics (IPC), Friedrich-Schiller-University, Jena, Germany
2
IPHT Working Group ’Statistical Modelling and Image Analysis’, Leibniz Institute of Photonic Technology (IPHT),
Jena, Germany
Keywords:
Non-linear Models, Taylor Series, Model Approximation, Model Interpretation.
Abstract:
Machine learning methods like classification and regression models are specific solutions for pattern recog-
nition problems. Subsequently, the patterns ’found’ by these methods can be used either in an exploration
manner or the model converts the patterns into discriminative values or regression predictions. In both ap-
plication scenarios it is important to visualize the data-basis of the model, because this unravels the patterns.
In case of linear classifiers or linear regression models the task is straight forward, because the model is
characterized by a vector which acts as variable weighting and can be visualized. For non-linear models the
visualization task is not solved yet and therefore these models act as ’black box’ systems. In this contribu-
tion we present a framework, which approximates a given trained parametric model (either classification or
regression model) by a series of polynomial models derived from a Taylor expansion of the original non-linear
model’s output function. These polynomial models can be visualized until the second order and subsequently
interpreted. This visualization opens the ways to understand the data basis of a trained non-linear model and
it allows estimating the degree of its non-linearity. By doing so the framework helps to understand non-linear
models used for pattern recognition tasks and unravel patterns these methods were using for their predictions.
1 INTRODUCTION
Pattern recognition is an emerging field, which finds
numerous applications in a wide range of other scien-
tific fields, e.g. in biology and chemistry (de S
´
a, 2001;
Bishop, 2011). In these both application fields the
aim is to extract useful information like patterns out
of high dimensional datasets from chemical and bio-
logical experiments (Bocklitz et al., 2014b). In these
application fields often supervised machine learning
methods like classification or regression approaches
are used to analyze un-targeted higher-dimensional
measurements, like images (Bocklitz et al., 2014a),
spectra (Kemmler et al., 2010; Bocklitz et al., 2009)
or time traces (Volna et al., 2016).
If the classification or regression task, which
should be solved, is complicated, non-linear models
are advisable. Because of the fact that biological or
chemical pattern recognition tasks are typically hard
to solve, non-linear models must be applied. Never-
theless, a drawback of these non-linear models is that
the patterns, which the model learned, cannot be ex-
tracted and visualized easily. In this way non-linear
classification and regression models work as ’black
box’ methods and no insight in their working wise
can be gained. This is a drastic drawback of these
non-linear methods, because in the application sci-
ence always an interpretation of the model is needed.
Such an interpretation would allow to gain new in-
sights into the classification task or regression task,
unravel pattern in the data and allow a check if the
model was learning artifacts.
To solve this interpretation task two schemes were
developed. One scheme is the calculation of variable
importance measures, like Gini importance or permu-
tation importance (Hapfelmeier et al., 2014), and the
other scheme is the visualization of instances, which
lead to an extreme prediction. Both workflows are
not optimal, because the variable importance do not
state, why and how a specific variable is utilized to
calculate quantitatively the output. Therefore, a direct
model interpretation approach is needed, which lead
to a direct understanding of the non-linear model. A
solution for this issue is presented in this article.
The outline of this contribution is as follows. In
section 2 the Taylor series and parametric models
874
Bocklitz, T.
Understanding of Non-linear Parametric Regression and Classification Models: A Taylor Series based Approach.
DOI: 10.5220/0007682008740880
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 874-880
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
like classification and regression techniques are intro-
duced. In section 3 our approximation scheme is pre-
sented and in section 4 the example datasets together
with the interpretation of the approximation models
with respect to the data are discussed. In section 5 the
paper is summarized and an outlook is given.
2 THEORETICAL
CONSIDERATIONS
2.1 Taylor Series
In this paragraph the Taylor series or Taylor expan-
sion is introduced. A Taylor series represents a given
function f : R 7→ R as an infinite sum of terms that can
be calculated in a small neighborhood around a given
point x
(0)
∈ R. These terms contain the values of the
function’s derivatives at the point x
(0)
and this point is
called expansion point. If the infinite sum is used and
it converges, the function and its Taylor series can be
used interchangeable
1
. If only finite terms of its Tay-
lor series are utilized an approximation of the function
f can be extracted. Then the Taylor’s theorem states
an estimate of the error such an approximation is fea-
turing. The polynomial formed by taking some initial
terms of the Taylor series is called a Taylor polyno-
mial and we will stick to this term through the arti-
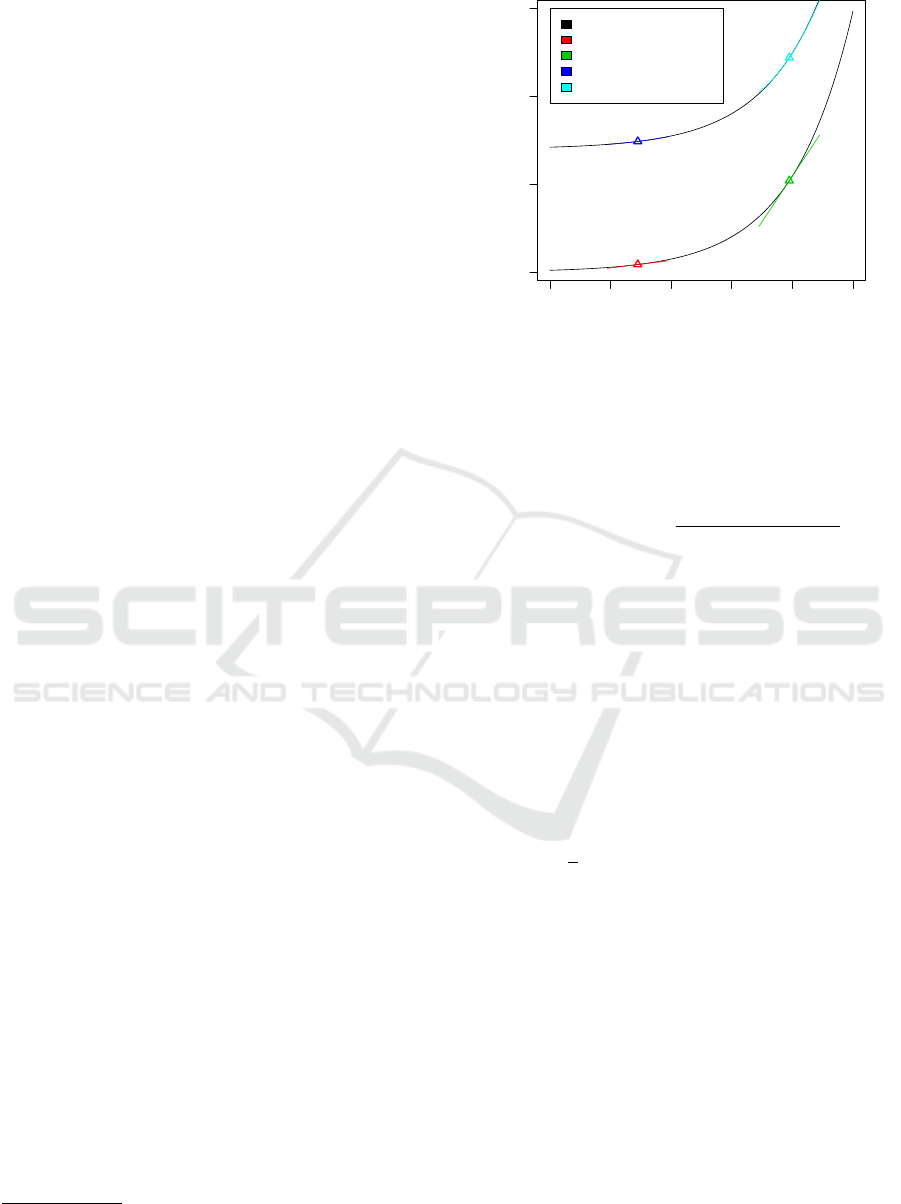
cle. In Figure 1 Taylor polynomials of two different
exponentials are given, which were calculated at two
different expansion points. In the upper exponential
second order polynomials are used for approximation,
while in the lower trace linear approximations were
tested. It is clear that the Taylor polynomials approxi-
mate the function in a neighborhood of the expansion
point x
(0)
quite well. The error of the approximation
can be calculated via different error formulas related
to the polynomials, which were not used in the ap-
proximation.
In the further cause of the article we need to work
with high dimensional functions, which map from the
R
n
to the real numbers and we will denote the cor-
responding function with F : R
n
7→ R. We will de-
note the points of the R
n
in bold: x ∈ R. The expan-
sion point is again termed x
(0)
∈ R. The Taylor series
can be calculated using the higher dimensional deriva-
tives, e.g. Gradient and Hessian. The corresponding
Taylor series (Bronstein et al., 2012) can be expressed
as:
1
The function needs to be an analytical function that this
equality holds true (Bronstein et al., 2012).
0 1 2 3 4 5
0 50 100 150
x
y
exponential increase
1. order approximation
1. order approximation
2. order approximation
2. order approximation
Figure 1: One-dimensional Taylor expansion. Linear and
quadratic approximations of two exponential functions in
two different expansion points are shown. In the neighbor-
hood of the expansion point the approximation quality is
appropriate.
F (x) = T
x
(0)
(x) =
∞
∑
i=0
x − x
(0)
i
· ∇
i
F|
x
(0)
i!
. (1)
If only the constant and linear term is used a Taylor
polynomial approximation results, which can be writ-
ten as follows:
F (x) ≈ T
(1)
x
(0)
(x) = F
x
(0)
+
x − x
(0)
· ∇F|
x
(0)
. (2)
If the quadratic term is added, a quadratic Taylor ap-
proximation is generated and can be written as:
F (x) ≈ T
(2)
x
(0)
(x) = T
(1)
x
(0)
(x)
+
1
2
x − x
(0)
T
· ∇
2
F|
x
(0)
·
x − x
(0)
. (3)
In this ways the function F can be approximated by
a quadratic function using only the values around the
expansion point x
(0)
.
2.2 Parametric Classification and
Regression Models
Two important groups of machine learning methods
used for pattern recognition tasks are classification
and regression models. Parametric models form an
important sup-group of these models and they learn
an output function, e.g. the parameters of this output
function, based on a given training dataset. Typically
this learning involves the solving of an optimization
problem in the training phase of the algorithm. After
Understanding of Non-linear Parametric Regression and Classification Models: A Taylor Series based Approach
875

this procedure is finished, the output function:
F : R
n
7→ R (4)
is fixed and can be used for prediction of new mea-
surements or instances. The prediction can be done
directly (in the case of regression models) or after the
application of a threshold to the output function to
form a class decision in case of classification models.
To derive different machine learning methods differ-
ent optimization problems are stated and in turn they
lead to different output functions. The output func-
tions have in common that certain parameters are cho-
sen beforehand (hyper-parameters) and other parame-
ters are optimized or estimated based on the trainings
dataset. Mathematically, this yields to the fact that
the output function is parametrized F (x;p,q). Some
of the parameters, e.g. the hyper-parameters q, are
fixed before training, while other parameters p have
to be determined in the trainings procedure based on
a given training dataset.
In the following, three examples of output func-
tions are given together with their parameters. The
output function of Fisher’s linear discriminant anal-
ysis (LDA) (Fisher, 1936), Vapnik’s support vector
machines (SVM) (Cortes and Vapnik, 1995) and ar-
tificial neural networks
2
(ANN) (Bishop, 1995) are
set together in the Equation 5 to 7. Due to its linear-
ity the output function of the LDA can be written as
scalar product with a learned vector s:
F (x) = (x · s) . (5)
The output of the LDA’s output function is subse-
quently converted via a threshold into a class predic-
tion. In contrast the output function of a SVM reads
as follows:
F (x) =
N
∑
i=1
y
i
α
i
K
x
(i)
,x
+ b
!
. (6)
While the number of support vectors i ∈
{
1,··· ,N
}
,
the coefficients α
i
and the value b are optimized, the
kernel function K is choose beforehand. Depending
on the chosen kernel K the SVM is a linear classifier
or a non-linear classifier. Another often applied re-
gression and classification model is the ANN and its
output function can be written as follows:
F (x) = f
0
"
n
H
∑
j=1
w
0
1 j
f
n
∑
i=1
w
ji
x
i
+ w
j0
!
+ w
0
10
#
. (7)
In this formula f , f
0
are the activation functions of
hidden and output layer. The number of neurons are
2
Here, we restrict ourselves to three layer feed forward
networks with one output neuron, but the generalization to
different topologies is straight forward.
n,n
H
. These values are the hyper-parameter, while the
weights w
ji
,w
0
1 j
and the biases w
0
10
,w
0
1 j
are learned.
These latter parameters are optimized while training
to minimize the summed error of all trainings in-
stances E =
∑
n
i=1
E
i
. The error of every individual
pattern is back propagated from layer to layer and
the derivatives of the error function with respect to
the weights and biases in the network is calculated
(Bishop, 1995; Rumelhart et al., 1986). Finally, the
weights and biases are updated accordingly.
3 PROPOSED METHOD
In order to get an insight into the patterns a non-
linear model is used for prediction, we combine the
aforementioned two concepts. Basically we generate
a quadratic approximation of the non-linear machine
learning model (regression or classification model),
which was initially used for pattern recognition. We
utilized a Taylor polynomial until the second order
(see Equation 3). If the linear part of the second order
Taylor polynomial is explicitly written, it looks like
x − x
(0)
· ∇F|
x
(0)
(8)
=
n
∑
i=1
x
i
− x
(0)
i
· ∇
F|
x
(0)
i
and can be understood as a variable weighting. The
value of the Gradient ∇F|
x
(0)
represents the weight
of the variables. The Equation 8 is similar to output
function of the LDA (Equation 5). A similar interpre-
tation as for the Gradient holds true for the quadratic
term and it reads (without the 1/2 factor)
x − x
(0)
T
∇
2
F|
x
(0)
x − x
(0)
(9)
=
n
∑
i=1
n
∑
j=1
x
i
− x
(0)
i
·
∇
2
F|
x
(0)
i, j
x
j
− x
(0)
j
.
The advantage of this approximation is that we can
quantify the degree of non-linearity of the original
non-linear model by comparing the prediction of the
original model with the predictions of approximations
with different degree. If the prediction of an approx-
imation is worse compared with the original model,
the non-linearity is at least higher as the approxima-
tion degree n. This fact results from the Taylor’s rest
formula, which state that the rest term is
O
x − x
(0)
(n+1)
. (10)
If the approximation performance in terms of the
model prediction, e.g. either accuracy or Root-Mean-
Squared-Error (RMSE), is acceptable, the approxima-
tion can be used instead of the original model. The
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
876
range of acceptable performance of the model can
only be discussed in terms of the application scenario
and is not further discussed here. If only a model up
to the second order is needed, e.g. its performance is
sufficient, the quadratic part and the linear part of the
model can be visualized and subsequently interpreted.
The visualization of the approximation can be done
by plotting the Gradient of the model, e.g. the lin-
ear part of the approximation, and the Hessian, e.g.
the quadratic part of the approximation, in false col-
ors. The Gradient can be interpreted due to Equation 2
and Equation 8 as a variable weighting. Additionally,
the sign and its magnitude can be interpreted, which
is an advantage over any variable importance score.
Beside this linear part of the approximation also the
quadratic part, e.g. the Hessian, can be visualized and
interpreted. Due to Equation 3 and Equation 9 the val-
ues of the Hessian on the main diagonal correspond
to pure quadratic dependencies of the output function
on the respective variable, while the off-diagonal ele-
ments belong to dependencies of the output function
on variable combinations. The magnitude and sign of
the Hessian values can be interpreted in a similar way
as above for the Gradient.
4 EXPERIMENTAL RESULTS
AND DISCUSSIONS
4.1 Datasets
We utilized two datasets respectively models to
demonstrate the developed approximation frame-
work. We used a low dimensional dataset for the
demonstration of the approximation of classification
models and a high dimensional dataset for the demon-
stration of the approximation of regression models.
The classification will be demonstrated on a sub-
set of Fisher’s Iris flower dataset (Fisher, 1936) and
we used the version shipped with the R package
’MASS’ (Venables and Ripley, 2002). This dataset
consist of 4 variables (the length and the width of the
sepals and petals, in centimeters) to discriminate three
species of Iris (Iris setosa, Iris virginica and Iris ver-
sicolor). We removed the species Iris setosa from the
dataset to form a binary classification task. We solved
this classification task using a 3-layer feed-forward
artificial neural network implemented using the ’nnet’
package (Venables and Ripley, 2002), which is called
Iris-ANN from here on. The hyper-parameters were a
quadratic error function, a linear output of the output
layer and there were 4 input neurons, 2 hidden neu-
rons and one output neuron. The group Iris virginica
5 10 15 20 25 30
5 10 15 20 25 30 35
x/µm
y/µm
−0.1
0.0
0.1
0.2
0.3
0.4
0.5
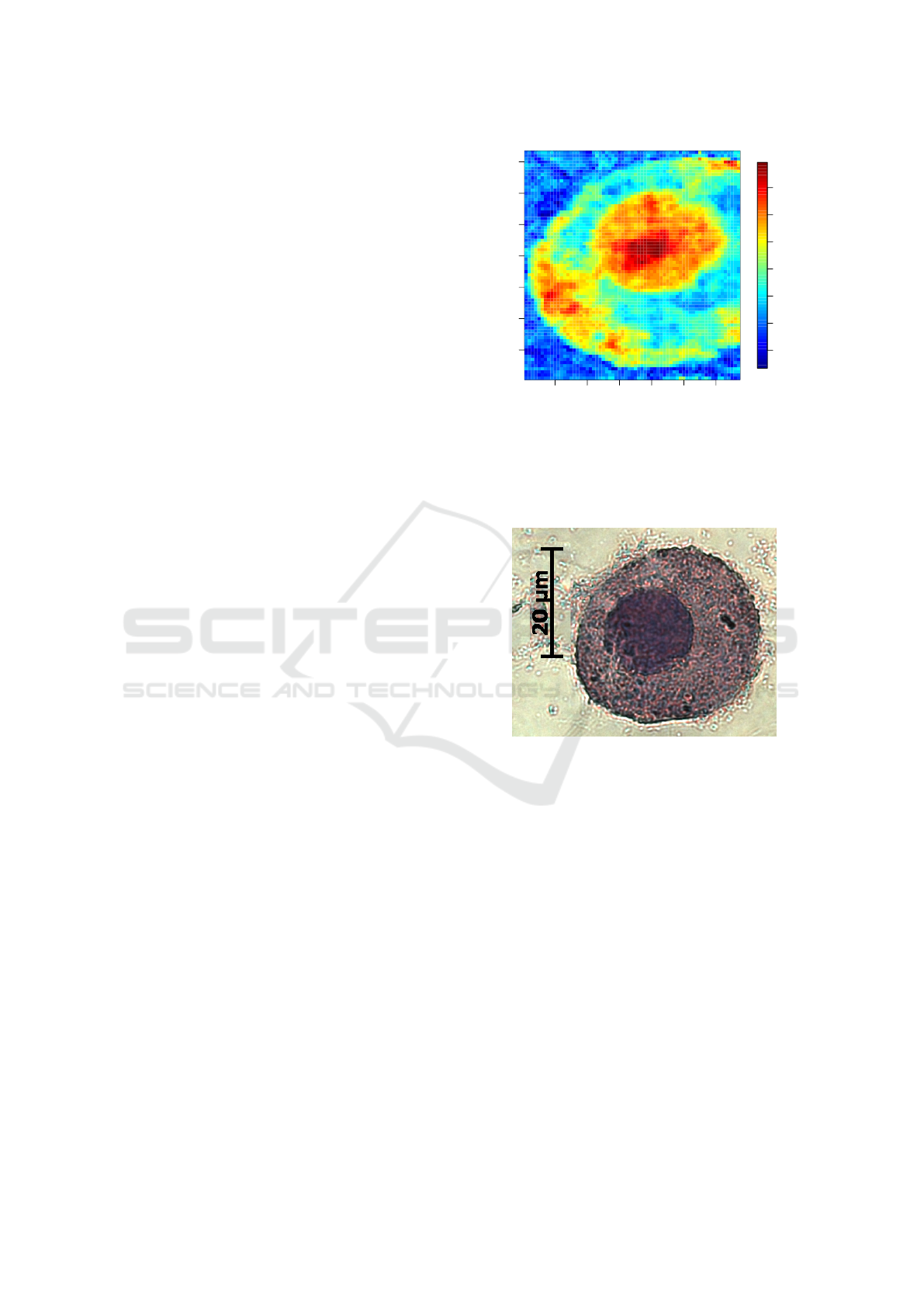
Figure 2: Output of the DNA-ANN for an examples cell.
The output of the DNA-ANN, which was trained based on
Raman spectral scans of cells, is shown. The visual ap-
pearance of the result indicated that the nucleus region is
highlighted.
Figure 3: HE image of an example cell. The staining of the
cell after Raman spectroscopic measurement was the only
validation of the DNA-ANN’s output.
was coded with 1, while the group Iris versicolor was
coded with 0.
The dataset and model used to demonstrate the
approximation of a regression model was published
in reference (Bocklitz et al., 2009). In this publica-
tion a ANN was trained to highlight the cell nucleus
and the model is called DNA-ANN from here on. It
was trained using Raman spectra of cells exhibiting a
strong DNA/RNA contribution. The model was using
a principal component analysis (PCA) for dimension
reduction to 60 PCs, which were used for training the
DNA-ANN. Ten hidden neurons and on linear out-
put neuron were utilized. The validation in reference
(Bocklitz et al., 2009) was done based on visual com-
parison of the output of the DNA-ANN (Figure 2)
with images of Hematoxylin and Eosin stained cells
(Figure 3). The question arose, which patterns in the
spectroscopic data, e.g. Raman bands, were used to
Understanding of Non-linear Parametric Regression and Classification Models: A Taylor Series based Approach
877

calculate the DNA-ANN output. With this informa-
tion the model could be understand and the Figure 2
can be interpreted accordingly.
4.2 Software
All computations were done in the free programming
environment R using the packages ’MASS’, ’nnet’
and ’numDeriv’.
4.3 Results – Classification Technique
In order to approximate a given classifier we first
trained an ANN based on the Iris dataset with only
one output neuron (Iris-ANN). The coefficients of the
approximation of the Iris-ANN are calculated using
Equation 3. Here the trained Iris-ANN was approxi-
mated by a second order Taylor polynomial T
(2)
x
(0)
(x),
which was expanded around the mean of the reduced
Iris dataset (see Equation 12). We also substitute the
difference x − x
(0)
by ∆x:
∆x = x − x
(0)
. (11)
For the reduced Iris data set this equation reads as fol-
lows:
∆x = x −
6.262
2.872
4.906
1.676
sepal length
sepal width
petal length
petal width
. (12)
In order to extract the Gradient and the Hessian from
the given non-linear model two possibilities arise.
First, the analytical derivatives or numerical estimates
of the derivatives can be utilized. Both methods are
suitable, but care has to be taken to include the dimen-
sional reduction, converting functions and/or scaling
steps, if the analytical formulas are used. The nu-
merical derivatives already include all three steps,
therefore we utilized the numerical approach (Gilbert,
2006).
The approximation of the Iris-ANN is called P
(2)
Iris
for
clarity reasons. The corresponding coefficients of the
approximation P
(2)
Iris
are given in Equation 13:
P
(2)
Iris
(∆x) = 0.46 +
−1.82
−3.12
5.17
6.29
· ∆x (13)
+
1
2
∆x
T
0.98 1.56 −2.54 −3.35
1.56 2.89 −4.26 −5.74
−2.54 −4.26 7.23 5.47
−3.35 −5.74 5.47 11.42
∆x .
The classification results on the training dataset are
summarized in Table 1 for the Iris-ANN and in Ta-
ble 2 for the approximation P
(2)
Iris
. The results of the
classifier P
(2)
Iris
(5 errors) are not as exact as the results
of the Iris-ANN (1 error), but the P
(2)
Iris
approximation
is interpretable and features only a low degree of non-
linearity. From the Gradient values (Equation 13) it is
clear that an increased sepal length and width is an in-
dicator for Iris versicolor, because it was coded with
zero in the trainings phase. Petal length and width
are linear important for the Iris virginica group. The
Hessian can be interpreted in the same manner. All
variables are quadratically linked to the Iris virginica
group, while combination of variables are negatively
correlated with the Iris virginica group. The large er-
ror rate might be attributed to a higher degree of non-
linearity of the Iris-ANN or that the Iris-ANN features
less parameter compared to the P
(2)
Iris
approximation.
Table 1: Confusion table of the Iris-ANN model.
predicted
classes true classes
versicolor virginica
versicolor 49 0
virginica 1 50
Table 2: Confusion table of the P
(2)
Iris
model.
predicted
classes true classes
versicolor virginica
versicolor 45 0
virginica 5 50
4.4 Results – Regression Technique
To prove the approximations and visualization ap-
proach for regression techniques, we also performed
a second order approximation of the described DNA-
ANN model published in (Bocklitz et al., 2009).
The expansion point was the dataset mean and we
termed the second order approximation P
(2)
DNA
. First,
we checked the quality of the approximation. To do
so, we compared the output of the DNA-ANN with
the output of the P
(2)
DNA
approximation. We visualized
both outputs in Figure 4. The output of both models
was sorted according to the DNA-ANN output, which
forms a sigmoidal shaped curve. The difference be-
tween both outputs is given in green at the bottom of
Figure 4. It can be seen at the edges around 0 and
around 8000, that only for extreme values a larger er-
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
878
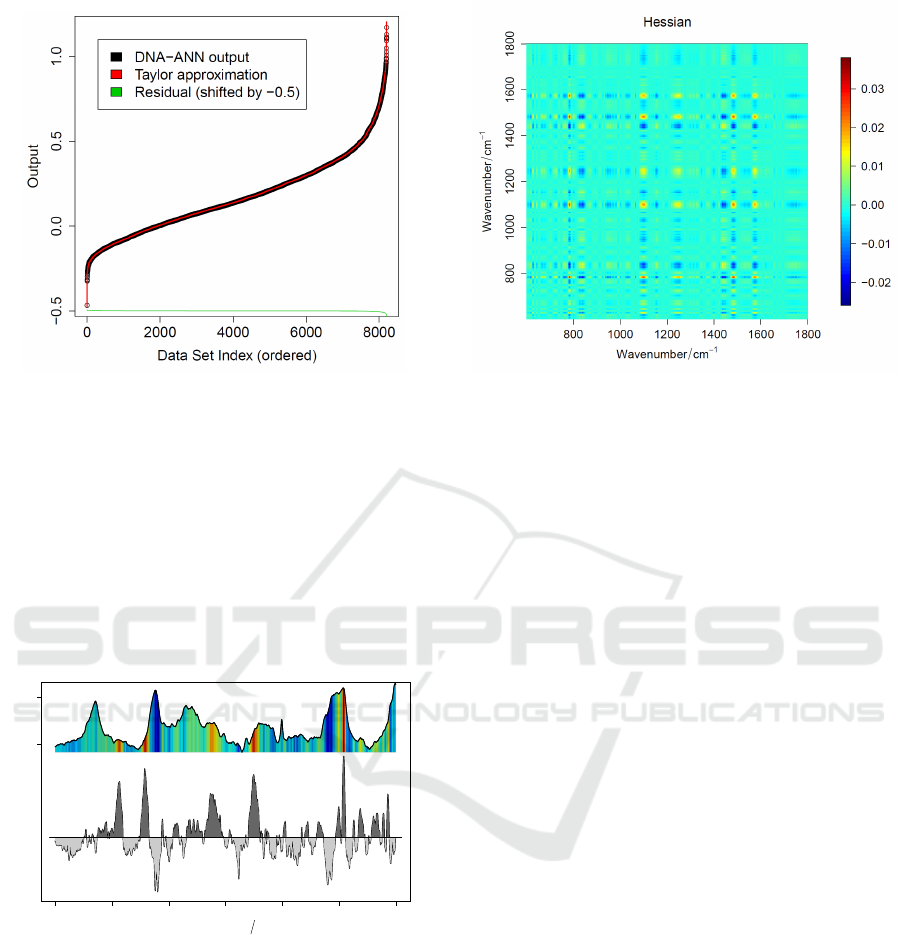
Figure 4: DNA-ANN output and approximation output.
The DNA-ANN output is visualized together with the out-
put of the P
(2)
DNA
approximation. In the lower part the differ-
ence of both outputs is plotted. The approximation quality
is good as the RMSE is only 0.0019.
ror between the DNA-ANN output and the P
(2)
DNA
out-
put exists. In this case the RMSE of the approxima-
tion was 0.0019. Therefore, the second order approx-
imation is sufficient to approximate the (given) DNA-
ANN model in an appropriate manner.
1800 1600 1400 1200 1000 800 600
Linear part of the approximation
Wavenumber cm
−1
Normalized Raman Intensity
0.01 0.06
−
+
Figure 5: Gradient ∇F|
−→
x
0
of the DNA-ANN and mean of
the dataset. The values of the Gradient are visualized in the
lower panel and they can be interpreted: positive features
in the Gradient mark areas which are connected quantita-
tively with a positive output of the DNA-ANN, e.g. a higher
DNA content. The spectrum in the upper panel represents
the dataset mean and the false-colors show the Gradient val-
ues.
Due to the low RMSE of only 0.0019, the
quadratic approximation can be used instead of the
original non-linear regression model. Because it is
composed of a linear and a quadratic part in a sum,
we can investigate the linear and quadratic behavior
separately. The linear part of the approximation is
Figure 6: Hessian ∇
2
F|
−→
x
0
of the DNA-ANN. Again the
positive areas mark variable combinations which are corre-
lating with a higher DNA-ANN output, while negative areas
mark the opposite.
plotted in Figure 5. This figure is composed of the
mean (upper panel) and the Gradient (lower panel).
The false color within the mean (upper panel) repre-
sents again the values of the Gradient. Positive values
in the Gradient can be interpreted that they are posi-
tively contributing to the model’s output with its value
as weight. Negative values are negatively used within
the output of the model. For example the sharp feature
at 785 cm
−1
marks a vibration of DNA/RNA and this
feature is positively correlated with the DNA-ANN’s
output, which indicate a correct interpretation of the
DNA-ANN.
The quadratic term can be also interpreted. The Hes-
sian is visualized in Figure 6 and its interpretation
can be done like in the case of the classifier above.
A positive or negative value on the main diagonal
means that the quadratic value of the corresponding
variable is positively or negatively connected with the
output of the DNA-ANN. This connection strength
is represented by the value itself. An off-diagonal
value is linked with variable combinations, which
then characterizes a positive or negative output, de-
pending on the sign (and magnitude) of the Hessian
value at the specific off-diagonal position. This in-
terpretation goes beyond variable/feature importance
measures, because the magnitude of the variable’s in-
fluence on the output is estimated and can be sub-
sequently interpreted. This interpretation possibility
leads to an understanding of the DNA-ANN.
Understanding of Non-linear Parametric Regression and Classification Models: A Taylor Series based Approach
879

5 CONCLUSION
In this contribution we presented a framework, which
allows the interpretation of patterns in the data, which
a parametric non-linear classification or regression
model was using for modeling. This framework is
working based on a Taylor expansion of the learned
output function of the respective model. This expan-
sion leads to a series of polynomial models (classi-
fiers, regressors), which can be used to understand
the non-linearity of a given non-linear model. A fur-
ther advantage of these polynomial approximations is
the fact that the linear and quadratic part can be vi-
sualized. By doing so, patterns in the data, which
were used in the modeling by the non-linear model,
can be elucidated. This approach can be used to ex-
tract, which variable or variable combinations are im-
portant to predict a special class or which are posi-
tively/negatively connected with the output of a re-
gression model. Nevertheless, the interpretation goes
beyond, because the magnitude of the variable influ-
ence on the output is estimated and can be interpreted.
This interpretation possibility is advancement over
variable/feature importance measures, which only in-
dicate important variables but not their specific, quan-
titative influence on the output. With this framework
non-linear models can be understood and they are not
working as ’black box’ systems anymore.
ACKNOWLEDGEMENTS
The funding of the Leibniz association via the
ScienceCampus ’InfectoOptics’ for the project
’BLOODi’, the funding of the DFG for the project
’BO 4700/1’ and funding of the BMBF for the
project URO-MDD (FKZ 03ZZ0444J) are highly
appreciated.
REFERENCES
Bishop, C. M. (1995). Neural Network for Pattern Recog-
nition. Clarendon Press.
Bishop, C. M. (2011). Pattern Recognition and Ma-
chine Learning. Information Science and Statistics.
Springer.
Bocklitz, T., K
¨
ammer, E., St
¨
ockel, S., Cialla-May, D., We-
ber, K., Zell, R., Deckert, V., and Popp, J. (2014a).
Single virus detection by means of atomic force mi-
croscopy in combination with advanced image analy-
sis. J. Struct. Biol., 188(1):30–38.
Bocklitz, T., Putsche, M., St
¨
uber, C., K
¨
as, J., Niendorf,
A., R
¨
osch, P., and Popp, J. (2009). A comprehensive
study of classification methods for medical diagnosis.
J. Raman Spectrosc., 40:1759–1765.
Bocklitz, T., Schmitt, M., and Popp, J. (2014b). Ex-vivo and
In-vivo Optical Molecular Pathology, chapter Image
Processing – Chemometric Approaches to Analyze
Optical Molecular Images, pages 215–248. Wiley-
VCH Verlag GmbH & Co. KGaA.
Bronstein, I. N., Hromkovic, J., Luderer, B., Schwarz, H.-
R., Blath, J., Schied, A., Dempe, S., Wanka, G., and
Gottwald, S. (2012). Taschenbuch der mathematik,
volume 1. Springer-Verlag.
Cortes, C. and Vapnik, V. (1995). Support-Vector Networks.
Machine Learning, 20(3):273–297.
de S
´
a, J. P. M. (2001). Pattern Recognition. Springer.
Fisher, R. A. (1936). The use of multiple measurements in
taxonomic problems. Annals Eugen., 7:179–188.
Gilbert, P. (2006). numDeriv: Accurate Numerical Deriva-
tives. R package version 2006.4-1.
Hapfelmeier, A., Hothorn, T., Ulm, K., and Strobl, C.
(2014). A new variable importance measure for ran-
dom forests with missing data. Statistics and Comput-
ing, 24(1):21–34.
Kemmler, M., Denzler, J., R
¨
osch, P., and Popp, J. (2010).
Classification of microorganisms via raman spec-
troscopy using gaussian processes. Pattern Recogni-
tion, online:81–90.
Rumelhart, D. E., Hinton, G. E., and Williams, R. J. (1986).
Learning representations by back-propagating errors.
Nature, 323:533–536.
Venables, W. N. and Ripley, B. D. (2002). Modern Applied
Statistics with S. Springer, New York, fourth edition.
Volna, E., Kotyrba, M., and Janosek, M. (2016). Pattern
Recognition and Classification in Time Series Data.
IGI Global.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
880
