
Genetic Diversity of Cowpea Mild Mottle Irus on Soybean in Several
Region in Indonesia
Mimi Sutrawati
1
, Sri Hendrastuti Hidayat
2
, Bonny Purnomo Wahyu Sukarno
2
, Gede Suastika
2
and
Ali Nurmansyah
2
1
University of Bengkulu, WR Supratman Street, Bengkulu, Indonesia
2
Bogor Agricultural University, Bogor, Indonesia
Keywords: DAS-ELISA, homology, nucleotide sequencing, PCR, phylogeny
Abstract: Soybean is one of the most important food commodities in Indonesia. Virus infection on soybean has been
reported worldwide as factors affecting yield loss. This study was aimed to detect Cowpea mild mottle virus
(CPMMV) from several soybean cultivation areas in Java, Sumatra and Southeast Sulawesi; and further
characterize their genetic variation based on nucleotide sequences of their coat protein. Several virus
infection was detected using double antibody sandwhich enzyme-linked immunosorbent assay (DAS-
ELISA),including CPMMV, Cucumber mosaic virus (CMV), and Soybean mosaic virus (SMV). Generally,
the symptoms caused by CPMMV, CMV, and SMV are similar, involving mottle, rugose, and vein banding.
Coat protein gene of 5 CPMMV isolates (Bantul, Musi Banyuasin, Cirebon, Kendari, Cianjur) was
successfully amplified and cloned. Sequence of this 5 clones of CPMMV showed high similarity, ranging
from 88.2 to 99.8%; whereas their sequence homology to those of Taiwan and China ranging from 88.2 to
98.6%. Phylogenetic analysis showed different clusters of CPMMV Indonesian isolates: isolates from
Bantul,Cirebon, Musi Banyuasin (Palembang) is clustered with Taiwan isolate (JX020701); isolate from
Cianjur is clustered with China isolate (KX534092); isolate from Kendari is clustered with Puerto Rico
(GU191840),Brazil (KC884247), and USA (KC774020) isolates.
1 INTRODUCTION
Several types of viruses reported to infect soybean
plants are Alfalfa mosaic virus (AlMV), Bean
common mosaic virus (BCMV), Bean yellow
mosaic virus (BYMV), Blackeye cowpea mosaic
virus (BlCMV), Cucumber mosaic virus (CMV),
Pea enation mosaic virus (PEMV), Peanut mottle
virus (PeMoV), Soybean mosaic virus (SMV),
Tobacco mosaic virus (TMV), Tobacco ringspot
virus (TRSV), Tobacco streak virus (TSV), Tomato
ringspot virus (ToRSV), and Spotted Tomato wilt
virus (TSWV) (Golnaraghi et al. 2004). In
Indonesia, several viruses have been reported in
soybean plants: Cowpea mild mottle virus
(CPMMV) (Iwaki et al. 1986), SMV (Andayanie
2012), CMV soybean strain (CMV-S) and Pepper
yellow leafcurl virus (PYLCV) ( Rahim et al. 2015).
CPMMV infection in soybean has caused endemic
diseases in Java and Sumatra (Jumanto et al. 1999).
CPMMV infection in soybeans in Lampung caused a
decrease in dry weight with soybean crop weight
between 15.5-53.4% and a decrease in soybean seed
weight between 11.5-51.6% and a decrease in seed
quality cause an abnormal seed shape of 7.6-54.35%
(Akin 2003).
CPMMV is a member of the Genus Carlavirus,
Family Betaflexiviridae (Martelli et al. 2007).
Losses due to CPMMV infection were also reported
in several other countries, including in Argentina
and Iran CPMMV reportedly caused severe damage
to soybeans (Laguna et al. 2006; Tavassoli et al.
2009). In addition to infecting soybean CPMMV
was also reported in yard long bean (Brito et al.
2012), tomatoes, beans, Bambara peanuts (Almeida
et al. (2005). Offei and Albrechtsen (2005) reported
CPMMV infection in bambara peanuts (Vigna
subterranea L.) causes the leaf area index to be
reduced by 70%, and decreases the number of pods
and seeds and the weight of the seeds CPMMV
infection in peanuts causes dwarf plants, reduction
in length of internodes and size of leaves, stripes on
leaves, chlorosis and leaf rolling CPMMV infection
has been reported to be a serious disease in peanuts
Sutrawati, M., Hidayat, S., Sukarno, B., Suastika, G. and Nurmansyah, A.
Genetic Diversity of Cowpea Mild Mottle Irus on Soybean in Several Region in Indonesia.
DOI: 10.5220/0009935718151819
In Proceedings of the 1st International Conference on Recent Innovations (ICRI 2018), pages 1815-1819
ISBN: 978-989-758-458-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
1815

in Sudan with a disease incidence of up to 50% even
in some regions up to 100% (El-Hassan et al. 1997).
Surveys on long bean plants in Venezuela show the
incidence of diseases ranging from CPMMV
infections. 15-40% (Brito et al. 2012). In India,
CPMMV infection in soybean causes systemic,
mosaic, and leaf deformation with 25.1-71 disease
incidence. % (Yadav et al. 2013).
Expansion of soybean cultivation areas in
Indonesia must anticipate the emergence of diseases
that have the potential to cause loss of results. The
current status of the area spread CPMMV on
soybeans in Indonesia needs to be known. Recently
on 2015, we conducted a field survey to collected
soybean’s leaves with typical symptom of virus-like
infection from several cultivation areas in Java,
Sumatra, and Sulawesi. In the earlier reports, the the
yield impact of CPMMV infection were studied, but
none of these were reported about genetic diversity
of CPMMV in Indonesia. Here, we reported the
genetic diversity of CPMMV from several soybean
cultivation areas in Indonesia.
2 MATERIAL AND METHODS
Sampling activities for collection of CPMMV
isolates were carried out during the 2014-2015
planting season in Cirebon, Cianjur, Bogor (West
Java Province); Bantul, (Yogyakarta); Ngawi (East
Java Province); Musi Banyuasin (South Sumatra
Province), Sungai Hitam (Bengkulu Province), Kota
Baru (Jambi Province); and Kendari (Southeast
Sulawesi Province). Virus detection and cloning
were carried out at the Plant Virology Laboratory,
Department of Plant Protection, Faculty of
Agriculture, IPB.
2.1 Samples Collection
Samples of soybean leaves taken from the field are
young leaves that show symptoms of viral infection
including mottle, vein clearing, chlorosis, dwarf,
leaves malformation. Leaf samples are put into a
plastic bag and stored in a box to be brought to the
laboratory. Symptoms of leafy leaves were then
separated into two groups, namely leaves with
symptomatic stripes, chlorosis, distortion of leaves
and dwarfs and a group of yellowing leaves. Each
sample is weighed 0.1 g each, labeled and stored at -
80 ˚C until it is used for the next stage.
2.2 Serological Detection
Serological detection of viruses was carried out with
DAS-ELISA following the Clark and Adams (1977)
protocol using 3 types of antiserum separately,
namely CMV, SMV, and CPMMV antiserum
(DSMZ, Germany).
2.3 Reverse Transcription-Polimerase
Chain Reaction (RT-PCR)
Total RNA extraction. Total RNA extraction was
carried out using RNeasy Plant Mini Kits (Qiagen,
Hilder, Germany) according to the Qiagen protocol
(Qiagen 2003). RNA extraction results were
synthesized into cDNA using reverse transcription
(RT) method. CDNA amplification was performed
using a specific primary pair for CP CPMMV gene.
The primary forward used was CPF (5'-
ATTAAGGATCCGAGTTGATTTAAATAAGT-3
') and the reverse CPR primer (5'-
ATTAAGAATTCCTTGTGATTGAAATTGCG-3')
with an expected amplification product measuring
958 bp. The composition of the PCR reactant
consisted of 12.5 µl Dream taq PCR master mix
(Thermo Scientific), 1 µl primer forward 10 µM, and
1 µl reverse primer 10 µM, 1 µl cDNA, and 9.51 µl
H2O. Amplification of cDNA with 94 ° C stages for
5 minutes, then followed by 35 cycles consisting of
denaturation of 94 ° C for 1 minute, annealing 45 °
C for 1 minute, and DNA synthesis (extension) 72 °
C for 2 minutes, then extension 72 ° C for 10
minutes and the cycle ends at 4 ° C. Amplification of
agarose gel follows the method described
previously. CPMMV isolates which showed a clear
and thick DNA band of ± 958 bp then continued to
cloning stage.
2.4 DNA Cloning and Sequences
Analysis
PCR products were cloned into pTZ57R/T aesy
vector system based on the protocol provided by
Thermo Scientific, USA. Plasmid DNA recombinant
was sequenced and analyzed. The nucleotide
sequences of the gene were aligned with those
corresponding virus sequences deposited in
GenBank database by using software Clustal-W
(www.ebi.ac.uk). Phylogenetic tree CPMMV was
constructed using the ClustalX Bio Edit version 7.05
program and the MEGA 5.0 program with the
neighbor-joining algorithm and 1000 repetition
bootstrap methods.
ICRI 2018 - International Conference Recent Innovation
1816

3 RESULT AND DISCUSSION
3.1 Results
3.1.1 Symptoms on Infected Soybean Leaves
Symptoms of viral infection in soybean in the field
varied, namely systemic chlorosis and leaf
distortion, systemic leaf and curly leaf surface,
systemic stripes with green patches, striped with
lamina blisters, stripes and leaf distortion, chlorosis
in leaf lamina and vein banding, and some mixed
symptoms in one plant (figure 1). The symptom
variation was depending on the soybean cultivar and
climate on each location.
Figure 1: Symptoms of viral infection in soybean:
systemic chlorosis and leaf distortion (A), systemic stripes
and curly leaf surface (B), systemic stripes with green
spots (C), striped with lamina of blister (D) leaves, stripes
and leaf distortion (E), and chlorosis in the leaf lamina and
vein banding (F).
3.1.2 Serological Test
Based on serological test using three antisera, there
are three virures detetcted on samples with various
frequences and showed multiple infection occurred
naturally (data not shown). CPMMV detected on
samples in 8 regions from 9 regions of origin of
soybean leaf samples. Besides CPMMV also
detected several other types of viruses, namely CMV
and SMV. CMV infection is detected in 4 regions,
namely Cirebon, Cianjur, Bantul, Kendari; while
SMV is only detected in Kendari and Cirebon.
CPMMV was consider as endemic disease on
soybean in Java, further characterization of
biological, physical and nucleic acid characters are
necessary to identify to confirm its existence in
Indonesia.
3.1.3 Amplification and Nucleotide
Sequences Analysis
CPMMV specific DNA was successfully amplified
from 5 CPMMV isolates from Bantul (B),
Banyuasin Musi (MB), Cirebon (CB), Kendari
(KN), and Cianjur (CR) (figure 2). Furthermore, the
PCR product is used for cloning stages.
Figure 2: DNA amplification visualization of CP CPMMV
gene from Bantul (B), Banyuasin Musi (MB), Cirebon
(CB), Kendari (KN), and Cianjur (CR), M, 1 kb DNA
(Thermo Scientific) marker.
Isolates from 5 regions in Indonesia have
homology between isolates as much as 88.2-99.8%
(Table 1). Similarity of nucleotide sequences
between isolates in Indonesia and Taiwan and China
isolates with a range of 88.2-98.6% higher than
similarities with isolates from America; while the
lowest homology is with Ghana isolates. CPMMV
isolates in Indonesia showed more than 72%
homology with all isolates compared to GenBank.
Table 1: The nucleotide sequences of CPMMV isolates in
Indonesia with sequences of isolates from several other
countries in GenBank.
Sequence comparison CP
gene
(%)
Among Indonesian isolates
88.2-
99.8
Between Indonesian isolates with Taiwa
n
and China
8.2-
98.6
Between Indonesian isolates with
Amerika
6.9-
88.8
Between Indonesian isolates with Ghana
6.9-
79.2
Genetic Diversity of Cowpea Mild Mottle Irus on Soybean in Several Region in Indonesia
1817
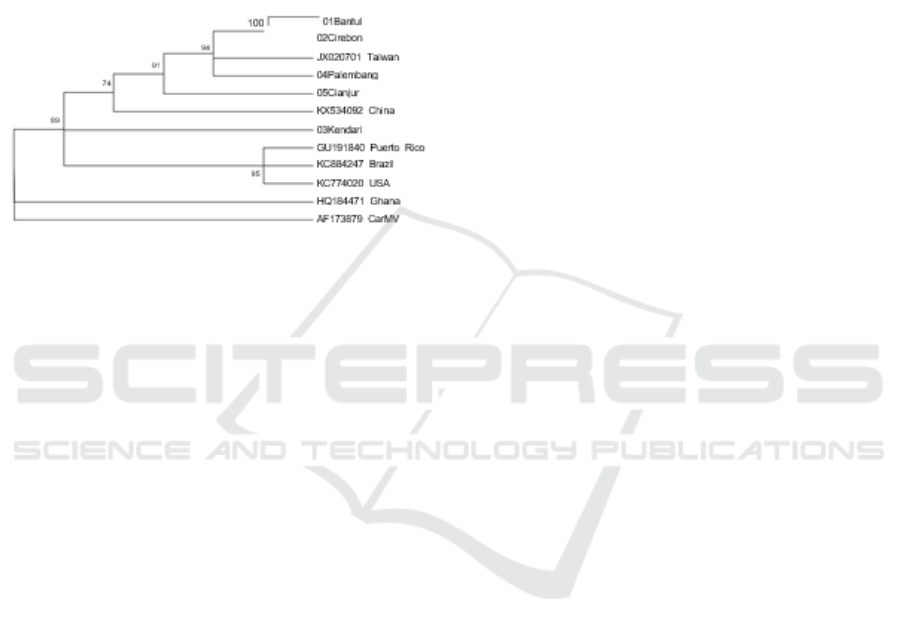
3.1.4 Phylogenetic Analysis
Phylogenetic analysis showed that Bantul, Cirebon,
Musi Banyu asin (Palembang) isolates formed a
group with Taiwan isolates (accession number
JX020701); whereas Cianjur isolates were separated
from other isolates and closer to Chinese isolates
(accession number KX534092). Kendari isolates are
closer to Puerto Rico isolates (accession number
GU191840, Brazil (accession number KC884247),
and USA (accession number KC774020) (Figure 3).
Figure 3. Phylogeny tree isolates CPMMV from Bantul,
Cirebon, Musi Banyuasin, Cianjur, and Kendari based on
the CP CPMMV gene sequence from Asian, American
and African groups and CarMV as outgroup
3.2 Discussion
Jumanto et al. (1999) reported mottled disease in
soybeans associated with CPMMV infection in Java
and Sumatra. However, after the report, there is no
current information regarding the distribution of
CPMMV in Indonesia, even though the area of
soybean cultivation in Indonesia is increasingly
widespread. In this study it was found that CPMMV
was detected on soybeans in Bengkulu, Musi
Banyuasin (South Sumatra); Cianjur, Bogor and
Cirebon in West Java; Bantul (Yogyakarta); Ngawi
(East Java); and Kendari (Southeast Sulawesi).
The results of this study indicate that the
diversity of CPMMV not only occurs in virulence and
the type of symptoms caused but also in diversity its
molecular level. Based on nucleotide homology
analysis, CPMMV isolates obtained from Sumatra,
Java, and Kendari in this study, had a relationship with
isolates from Asia (China, Taiwan), and America
(USA, Brazil, Puerto Rico). Close relationship with
Asian and American isolates indicates the possibility
of CPMMV Indonesia coming from that country.
Nevertheless, CPMMV from various regions turned
out to show genetic variation at the level of its
nucleotide sequence. Genetic variation in viruses can
occur through two events, namely mutation and
recombination. The high rate of mutation in the RNA
virus indicates an evolutionary strategy. According to
Agrios (2005) the evolution of viruses occurs as a form
of adaptation to environmental suitability, such as host
plants, strains of viruses, insect vectors. Different
environmental conditions between regions in
Indonesia, and climate differences between Indonesia
and other countries may cause environmental stresses
that cause genetic changes in the virus.
4 CONCLUSIONS
The distribution of CPMMV on soybeans covering
several areas of soybean production field in Java,
Sumatra, and Sulawesi. CPMMV is the dominant
virus found in 8 soybean cultivation locations from 9
sampling locations. Besides CPMMV, CMV, and
SMV were also found. Based on phylogenetic
analysis CPMMV Bantul, Cirebon, and Musi
Banyuasin isolates formed a group with Taiwan
isolates; whereas Cianjur isolates are closer to
Chinese isolates; and Kendari isolates are closer to
isolates from Puerto Rico, Brazil, and USA.
ACKNOWLEDGEMENTS
If any, should be placed before the references
section without numbering.
REFERENCES
Agrios, G.N., 2005. Plant Pathology, San Diego (US).
Academic Press.
Akin, H.M., 2003. Respon beberapa genotipe kedelai
terhadap infeksi cowpea mild mottle virus. J.
Hama dan Penyakit Tumbuhan Tropika. 3(2):40-
43.
Andayanie, W.R., 2012. Diagnosis penyakit mosaik
(Soybean mosaic virus) terbawa benih kedelai.
Jurnal Hama dan Penyakit Tumbuhan Tropika.
12(2).
Brito, M., Fernandez-Rodriguez T, Garrido MJ, Mejias A,
Romano M, Marys E. 2012. First report of
Cowpea mild mottle Carlavirus on yardlong bean
(Vigna unguiculata subsp. sesquipedalis) in
Venezuela. Viruses. 4(12):3804-
11. doi:10.3390/v4123804.
Clark, M.F., Adams, A., 1977. Characteristics of the
microplate method of enzyme-linked
immunosorbent assay for the detection of plant
viruses. J Gen Virol. 34(3):475-483.
El‐Hassan, S., Naidu, R., Ahmed, A., Murant, A., 1997. A
serious disease of groundnut caused by Cowpea
mild mottle virus in the Sudan. J of Phytopathol.
145(7):301-304.
ICRI 2018 - International Conference Recent Innovation
1818

Golnaraghi, A., Shahraeen, N., Pourrahim, R., Farzadfar,
S., Ghasemi, A., 2004. Occurrence and relative
incidence of viruses infecting soybeans in Iran.
Plant Dis. 88(10):1069-1074.
Iwaki, M., Thongmeearkom, P., Honda, Y., Prommin, M.,
Deema, N., Hibi, T., Iizuka N., Ong, C., Saleh, N.,
1986. Cowpea mild mottle virus occurring on
soybean and peanut in southest Asian countries.
Tropical Agriculture Research Centre, Tech. Bull.
(21):106-120.
Jumanto, H., Roechan, M., Muhsin, M., Asadi, M.N.,
Sawahata, H., 1999. Distribution of soybean virus
diseases in Indonesia. Risearch Institute for Food
Crop Biotechnology, Bogor.
Laguna, I.G., Arneodo, J.D., Rodríguez-Pardina, P.,
Fiorona, M., 2006. Cowpea mild mottle virus
infecting soybean crops in northwestern
Argentina. Fitopatologia brasileira. 31(3):317-
317doi:http://dx.doi.org/10.1590/S0100-
41582006000300015
Martelli, G.P., Adams, M.J., Kreuze, J.F., Dolja, V.V.,
2007. Family Flexiviridae: a case study in virion
and genome plasticity. Annu Rev Phytopathol.
45:73-100.
doi:10.1146/annurev.phyto.45.062806.094401.
Rahim, Y.F., Damayanti, T.A., Ghulamahdi, M., 2015.
Deteksi virus yang menginfeksi kedelai di Jawa.
Jurnal Fitopatologi Indonesia. 11(2):68.
Tavasoli, M., Shahraeen, N., Ghorbani, S., 2009.
Serological and RT-PCR detection of Cowpea
mild mottle carlavirus infecting soybean. Journal
of General and Molecular Virology. 1(1):007-011.
Yadav, M.K., Biswas, K.K., Lal, S.K., Baranwal, V.K.,
Jain, RK., 2013. A Distinct Strain of Cowpea mild
mottle virus Infecting Soybean in India. J
Phytopathol. 161(10):739-744.
doi:10.1111/jph.12119.
Genetic Diversity of Cowpea Mild Mottle Irus on Soybean in Several Region in Indonesia
1819
