
Degradation of Di-N-Butyl Phthalate by Microbacterium Aoyamense
Atpm-11 Isolated from Waste Water Treatment Plant
Ke Zhang
*
, Wei Chen and Jia Chen
College of Civil Engineering, Sichuan Agricultural University, Dujiangyan,Sichuan, 611830, China
Email: zhangke@sicau.edu.cn
Keywords: Di-n-butyl phthalate (DBP), microbacterium aoyamense ATPM-11, biodegradation, characteristics
Abstract: An efficient di-n-butyl phthalate bacterial strain ATPM-11 was isolated from activated sludge of waste water
treatment plant (WWTP). Based on its morphological, physiobiochemical characteristics and 16S rRNA
gene sequence, strain ATPM-11 was identified as Microbacterium aoyamense sp. The degradation
characteristics were investigated under different environmental conditions. The results showed that the
optimal temperature and pH for DBP degradation by ATPM-11 was 25 ℃ and 8.0, respectively. Under
these conditions, ATPM-11 could effectively degraded more than 83% of DBP at 400 mg/L. The diversity
of degradable substrates showed that strain ATPM-11 could degrade phthalate (DMP), DEP and DOP
efficiently. Therefore, this bacterial strain has potential to be used in DBP bioremediation.
1 INTRODUCTION
Phthalic acid esters (PAEs), one of the synthetic
organic compounds, are the wildly used and a higher
productivity as plasticizers, adhesive, additives,
paint solvent and Printing inks in the world (Li et al.,
2005). However, with the broad use of plastic
products, the phthalic acid esters abound in the
environment and they can migrate into the soil and
rainwater, thus enter the water system, which may
harm aquatic organisms and human health (Bai et al.,
2012). Di-methyl phthalate (DMP), di-n-butyl
phthalate (DBP) and di-n-octyl phthalate (DOP)
have been listed as priority pollutants by China
National Environmental Monitoring Center and the
US Environmental Protection Agency (Wang et al.,
2008). PAEs can be degraded by chemical and
physical methods, but microbial technology was
regarded as the most efficient way duo to it high
efficiency and low toxicity (Wan, 2012). The
hydrolysis and photolysis of DBP in the natural
environment are very slow and are difficult to
degrade. The physical method mainly consists of
humic acid or activated carbon adsorption, relying
on the strong pore structure and adsorption capacity
of adsorbent to remove DBP in water (Li et al.,
2013). The chemical method is mainly
photocatalytic degradation, which is the removal of
DBP in water by ultraviolet light. Although physical
and chemical methods have a good effect on the
removal of DBP in water body, there are obvious
defects, such as the final destination of DBP
attached to the adsorbent. In comparison, the
biological method is low cost and high efficient.
(Guo et al., 2007; Ding, 2012; Zheng et al., 2007)
Presently, several PAEs-degrading bacterial
strains belonging to the Gordonia sp.( Sarkar et al.,
2013), Enterobacter sp.( Fang et al., 2010) and
Arthrobacter sp.( Wen et al., 2014). They can be
isolated from different environments, while their
degrading efficiencies in other PAEs were low and
far from meeting the actual pollution control
requirements.Therefore, in order to improve the
biodegradation rate of phthalate esters, it’s necessary
to isolate highly effective degradation bacteria(Li et
al., 2014).
In the study, a DBP-degrading bacterium was
isolated from active sludge and identified by 16S
rDNA sequence. The biodegradation kinetics and
different environmental factors affecting this process
were investigated.And this study is expected to
improve current understanding of the bioremediation
of DBP and find higher effective DBP-dergading
strains.
Zhang, K., Chen, W. and Chen, J.
Degradation of Di-N-Butyl Phthalate by Microbacterium Aoyamense Atpm-11 Isolated from Waste Water Treatment Plant.
In Proceedings of the International Workshop on Environment and Geoscience (IWEG 2018), pages 11-15
ISBN: 978-989-758-342-1
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
11

2 MATERIALS AND METHODS
2.1 Reagents and Chemicals
DBP (99.5% purity) for the experiment was
purchased from Chengdu Kelong Chemical Reagent
Co., Ltd., All the chemical reagents were of
analytical grade and all solvents(Ethyl acetate and
methanol) were of HPLC grade purchased from
Tianjing kemiou Reagent Co., Ltd China. The
minimal medium (MM) contained (1L): MgSO
4
7H2O 0.5 g, K
2
HPO
4
1.70 g, FeSO
4•
7H
2
O 0.05 g,
and NaNO
3
0.5 g, (NH
4
)
2
SO
4
1.0 g,Na
2
MoO
4
0.0024
g, CaCl
2•
2H
2
O 0.04 g,FeCl
3
0.0018 g.The nutrient
broth (NB) for bacteria enrichment consisted of beef
extract 3g, peptone 5 g, NaCl 5 g, pH 7.2.Nutrient
agar plates were made using Nutrient Broth (NB)
supplemented with 2% agar.
2.2 Enrichment and Isolation of Dbp
Strains
The enrichment procedure was according to Wu
(Wu et al., 2010) with some modifications. Initially,
5.0 g of sludge was added to a 500-ml Erlenmeyer
flask containing 200 ml of MM solution amended
with concentration of 100 mg/l DBP. The
suspension was incubated for 6 days in the dark at
25 ℃according to pre-experiment on a rotary shaker
operated at 140 rpm. Subsequently, 2ml of the
enrichment culture was serially transferred five
times to fresh medium incubated under the same
conditions. At the same time, in the process of
transfer, containing a higher concentration of DBP
200–500 mg/L each time. Then the final enrichment
was streaked onto MM agar plates supplemented
with a mixture of DBP (500 mg/L) and incubated 1
week at 25℃. Presumptive colonies were picked on
the basis of differences in colony morphology and
coloration and re-streaked onto MM agar plates
amended with DBP. The bacterial isolates were
further purified by streaking on Nutrient Agar plates
and then re-streaked onto MM agar plates with and
without DBP to confirm their degradation abilities.
Isolates can grow in the presence of DBP but not in
their absence were selected for further study.
2.3 Amplification of 16S rDNA
Extraction kit (Sangon Corporation, Shanghai,
China) was used for the extraction of bacterial
genomic DNA according to the manufacturer’s
instructions. Further identification was performed by
16S rDNA gene sequencing. and then about 1500 bp
length of 16S rRNA was amplified through PCR by
using the bacterial universal primer 27F (50-
AGAGTTTGATCCTGGCTCAG-30) and 1492R
(50-GGCTACCTTGTTACGACTT-30). PCR was
performed ( Bio-Rad USA) under the following
conditions: preheated at 95 ℃ for 2 min; then
denatured at 94 ℃ for 1 min, annealing at 56 ℃ for
1 min, extended at 72 ℃ for 3min for 30 cycles, last
extended at 72 ℃ for 8 min.
2.4 Sequence Analysis of Strain
Purified PCR product was directly sequenced. The
sequence data of the closest relatives were retrieved
from NCBI database and aligned with CLUSTALW
with all parameters set at their default values. A
phylogenetic tree was then constructed using the
neighbor-joining method with MEGA 6.0 software.
The trees were validated using bootstrap analysis
performed with 1000 replicates.
2.5 Degradation Experiments of
Microbacterium Aoyamense
ATPM-11
The following environmental factors were assayed
to investigate their effects on DBP degradation
within 60 h of cultivation at a 140 rpm shaking rate.
Temperature (10, 15, 20, 25 and 30 ℃); Initial pH
value (4.0, 5.0, 6.0, 7.0, 8.0, 9.0); Initial DBP
concentration (100 mg/L, 200 mg/L,300 mg/L,400
mg/L,500 mg/L). Other PAEs (DOP, DEP, DMP,
DEHP and DPP).
IWEG 2018 - International Workshop on Environment and Geoscience
12

Figure 1: Phylogenetic tree derived from 16S rRNA gene sequence of ATPM-11 and sequences of related species.
Distances were calculated using neighbor-joining method.
2.6 Analysis Method
Concentration of DBP in the supernatant solution
was performed using high performance liquid
chromatography (HPLC) (Aglient 1200 series). The
column temperature was 40℃. The volume of the
injected samples was 40μl; Chromatography column
was Inertsil ODS-2151-K. 6× 150 mm. Ethyl acetate
was added to each sample, which was vigorously
shaken for 5 min, the aqueous and organic phases
were separated by centrifugation at 8000 rpm for 5
min. The aqueous phase was extracted twice with
equal volume of ethyl acetate. Ethyl acetate phase
was dried over anhydrous sodium sulfate and
evaporated, then dissolved in 10 ml of methanol.
3
RESULTS AND DISCUSSION
3.1 Isolation and Identification of the
DBP-degrading Bacterium
After 35 days enrichment, one strain showed high
biomass and high degradation efficiency was
selected for further investigation. Phylogenetic tree
of the 16Sr RNA gene revealed strain
Microbacterium aoyamense ATPM-11 clustered
with members of the genus Microbacterium, and
had a 100% sequence similarity with
Microbacterium aoyamense JCM 14900 (AB234028)
(Figure 1).
3.2 Effects of Temperature on DBP
Biodegradation
The strain was cultivated under condition of 25℃
and pH 8. The DBP (500 mg/L) degradation
efficiencies under 10, 15, 20,25and 30 ℃ were
tested. The effects of temperature on the degradation
of DBP in the culture medium were tested after
incubation 60 h. The results indicate that the
degradation is best at the temperature of 25 ℃, and
The degradation rate was 83% (Figure 2). The
degradation rate was only 30%. There is no
significant difference between 20 ℃ and 25℃
(P<0.05).
10 15 20 25 30
30
40
50
60
70
80
90
degradation(%)
Tem perature(
C)
Figure 2: Effects of temperature on DBP biodegradation.
3.3 Effects of Initial pH on DBP
Biodegradation
Figure 3 shows the results of pH (4.0–9.0) on DBP
biodegradation at an initial concentration of DBP at
400 mg/L. Strain could effectively degraded DBP
when pH ranged from 7.0 to 9.0. The optimal
Degradation of Di-N-Butyl Phthalate by Microbacterium Aoyamense Atpm-11 Isolated from Waste Water Treatment Plant
13
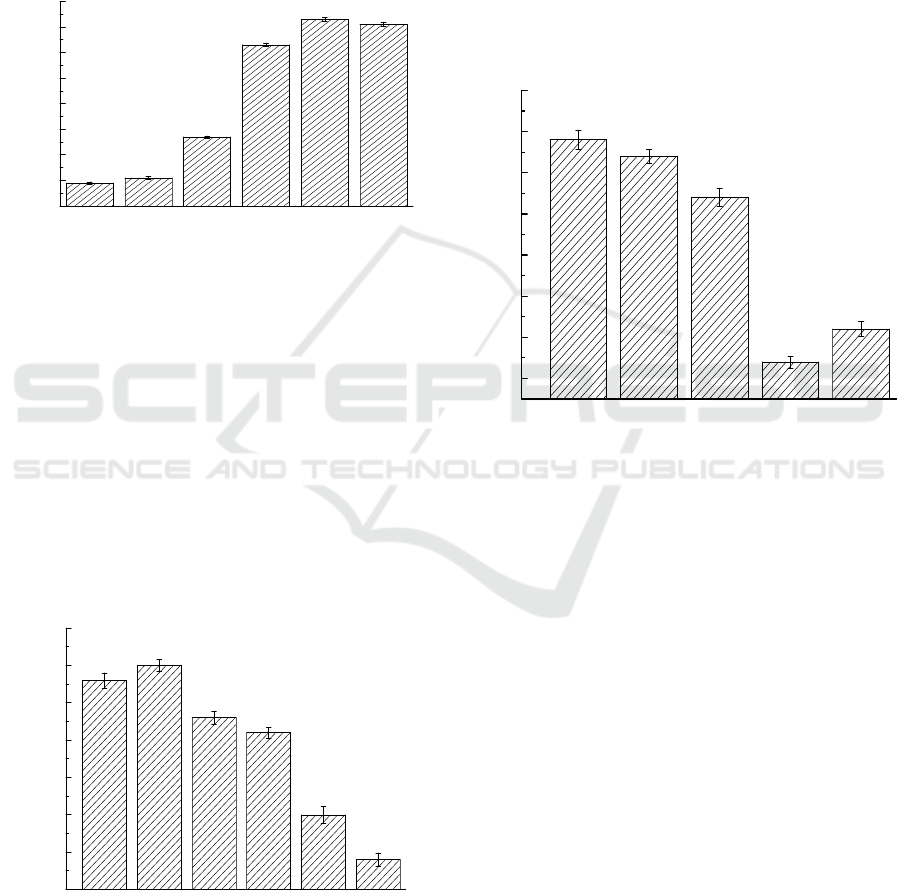
degradation pH for DBP degradation by this
bacterial strain were 8.0. The degradation rate could
reach up to 93%. The degradation under acid
conditions is poor. The optimal pH values in
degrading of DBP are consistent with the other
study, where the optimal pH for Di-n-butyl phthalate
(DBP) Degradation by strain H-2 ranged from 7.0 to
9.0. (Lei et al., 2014).
456789
20
30
40
50
60
70
80
90
100
Degradation rate (%)
pH
Figure 3: Effects of initial pH on DBP biodegradation.
3.4 Effects of Initial Concentration on
DBP Biodegradation
The experiment was conducted under different DBP
concentrations (100,200,300,400,500,600 mg/L) to
investigate the influence of concentration on DBP
degradation rate. As shown in Figure 4, when the
initial concentration was 200 mg/L, Microbacterium
aoyamense ATPM-11 had the highest degradation
rate of DBP. The degradation rate was slightly lower
than 200 mg/L when the concentration was 100mg/L.
when the concentration gradually increased from
200mg/L to 600mg/L, the degradation rate reached
the lowest value of 69%.
100 200 300 400 500 600
65
70
75
80
85
90
95
100
Degradation rate (%)
Concentration(mg/L)
Figure 4: Effects of initial concentration on DBP
biodegradation.
3.5 Degradation of Other PAEs by
Bacteria
In order to investigate the degradation ability of the
consortium to other commonly used PAEs in
environment, the consortium was cultured in MSM
supplemented with DBP, DOP, DEP, DMP, DEHP
and DPP at 30 °C. Figure 5 shows that
Microbacterium aoyamense ATPM-11 can also
degrade the other APEs. The strain could degraded
DEP and DMP with high efficiency up to 94 %.
DEP DMP DOP DEHP DPP
65
70
75
80
85
90
95
100
Degradation rate (%)
PAEs
Figure 5: Degradation of other commonly used PAEs by
bacteria.
4 CONCLUSIONS
A broad-spectrum and efficient di-n-butyl phthalate
(DBP)-degrading bacterial strain Microbacterium
aoyamense ATPM-11 was isolated from activated
sludge of waste water treatment plant (WWTP). The
strain Microbacterium aoyamense ATPM-11 could
completely degrade DBP and the degradation rate
was up to 93%. The temperature 25 ℃ and pH 8.0
are the optimal conditions for DBP degradation by
strain ATPM-11. Microbacterium aoyamense
ATPM-11 degrades DBP faster in alkalinity than in
acidity. The strain Microbacterium aoyamense
ATPM-11could also degrade other commonly used
phthalates like (DMP), DEP and DOP.
IWEG 2018 - International Workshop on Environment and Geoscience
14

REFERENCES
Bai YH, Xu K and Zhao DB 2012 Exposure assessment of
Di (2-ethylhexyl) phthalate of plastic food packaging
materials. Modern Food Science and Technology
28(10) 1423-1428
Ding K 2012 Experimental research on carbon for the
adsorption of DBP from water and dielectric barrier
discharge [D]. Nanchang: Nanchang University
Fang CR, Yao J, Zheng YG, Hu LF and Jiang CJ 2010
Dibutyl phthalate degrationby Enterobacter sp. T5
isolated from municipal solid waste inland fill
bioreactor. International Biodeterioration and
Biodegradation 64(6) 442-446
Guo D, Li Y, Zhao Y and Zhou J 2007 Activated carbon
adsorption of di-n-butyl phthalate, nonyl phenol and
bisphenol A in Yellow River water. Water Wastewater
Eng 33(1) 30-33
Li JL, Shao XL , Liu SL, Yao K, Zhao Q, Hu XJ and
Deng WQ 2014 Screening and identification of
dibutyl phthalate degradation strains. Modern Food
Science and Technology 30(10) 10-113
Li JX, Gu JD and Li P 2005 Transformation of dimethyl
phthalate, dimethyl isophthalate and dimethyl tereph
Li thalate by Rhodococcus rubber Sa and modeling
the process using themodified gompertz model.
International Biodeterioration and Biodegradation
55(3) 223-232
Li YH,Yi YC,Wu J, Wu J and Na W 2013 Isolation of a
humus-reducing bacterium strain and its
biodegradation of pollutants [J]. ChinJ Appl Environ
Biol 19(3) 506-510
Lei J, Chen Y, Long J, Guo YM, Yan ZY,Zhu Y and Gu
PQ 2014 Isolation and Identification of a Di-n-butyl
phthalate (DBP)-Degrading Strain H-2 and Its
Degradation Characteristics. Journal of Food Science
35(15) 203-206
Sarkar J, Chowdhury P P and Dutta T K 2013 Complete
degradation ofdi-n-octyl phthalate by Gordonia sp.
strain Dop5 [J].Chemosphere 90 2571-2577
Wan Y 2012 Study on Adsorption-desorption
characteristics of dibutyl-phthalate on humic acid [D].
Chongqing: Southwestern University
Wang P, Wang S and Fan CQ 2008 Atmospheric
distribution of particulate- and gas-phase phthalic
esters (PAEs) in a metropolitan city, Nanjing, East
China, Chemosphere 12 (72) 1567–1572
Wen ZD, Gao DW and Wu WM 2014 Biodegradation and
kineticanalysis of phthalates by an Arthrobacter strain
isolated from constructed wetland soil. Applied
Microbiology and Biotechnology 1-8
Wu XL, Liang RX, Dai QY, Jin DC, Wang YY and Chao
WL 2010 Complete degradation of di-n-octyl
phthalate by biochemical cooperation between
Gordonia sp. strain JDC-2 and Arthrobacter sp. strain
JDC-32 isolated from activated sludge. Journal of
Hazardous Materials 176 262–268
Zheng HH, Zhao LW and Tao J 2007 Photo degradation
of dibutyl phthalate in flowing water. Chin J Public
Health 23(2) 210-211
Degradation of Di-N-Butyl Phthalate by Microbacterium Aoyamense Atpm-11 Isolated from Waste Water Treatment Plant
15
