
Distribution and Phylogenetic Diversity of CbbL Gene Encoding
RuBisCo in the Deep-sea Sediments from the South China Sea
Jie Su
1
, Hongxia Ming
1
, Quanrui Chen
1,2
, Yuan Jin
1,3
, Chunxin Zhang
1,2
, Daoming Guan
1
and
Jingfeng Fan
1,3*
1
Key Laboratory for Ecological Environmental in Coastal Areas(SOA), National Marine Environmental Monitoring
Center, Dalian China 116023;
2
College of Fisheries and Life Science, Dalian Ocean University, Dalian China116023;
3
Fourth Institute of Oceanography, State Oceanic Administration, Beihai China 536000
Email: jffan@nmemc.org.cn
Keywords: Phylogenetic diversity, deep-sea sediments, South China Sea
Abstract: RuBisCO is the key enzyme catalyzing the first and major step of carbon fixation in Calvin cycle. With the
aim to examine the phylogenetic diversity of RuBisCO genes, cbbL gene was amplified by PCR from three
South China Sea deep-sea sample, cloned, and sequenced. A total of 28 OTUs deriving from 236 cbbL
clones covered 16 phylotypes which all belong to Proteobacteria. The estimated coverage values showed
that more than 85% of bacterial cbbL diversity was captured. Shannon and Simpson indices indicated a high
diversity of the total cbbL gene in the SCS deep-sea sediments. In conclusion, microbial RuBisCO genes in
the South China Sea display a broad range of phylogenetic diversity. The predominant group in these three
deep-sea sediments included Thiobacillus and Thiorhodococcus, which was chemoautotrophic bacteria
involving in Calvin–Benson–Bassham cycle. The above results implicate that bacteria with the potential for
carbon dioxide fixation and chemoautotrophy oocur in the South China Sea.
1 INTRODUCTION
CO
2
is the major contributor to global warming and
reduction of CO
2
input in atmosphere is essential for
control global warming. The ocean is recognized as
a huge carbon reservoir. Deep-sea sediments are a
huge carbon pool, the study of microbiology in
deep-sea ecosystem mediating flows of energy in
metabolism will help understand the CO
2
fixation
capacity of the marine ecosystem (Coffin, 2004).
Biologically mediated CO
2
fixation is a major
pathway in marine ecosystem. Microbiological
auotrophic CO
2
fixation is widely distributed and
adapted to many habitats.
Autotrophic CO
2
fixing bacteria do not belong to
a specific taxonomic group and occurs in many
species. Most chemolithoautotrophic bacteria
mediate autotrophic CO
2
fixation via the Calvin–
Benson–Bassham cycle (Shively, et al., 1986; Selesi
et al., 2005) RuBisCO is the key enzyme catalyzing
the first and major step of carbon fixation in the
Calvin cycle and exists in multiple natural forms
which differ in structure, catalytic property, and O2
sensitivity. As such, RuBisCO form I-encoding
cbbL genes have been used as functional markers for
molecular ecological studies of CO
2
assimilative
autotrophs in aquatic systems (Yuan et al., 2012;
Kovaleva et al., 2011).
The South China Sea (SCS), near to the West
Pacific “warm pool” is the biggest and deepest sea
in China (Dai et al, 2002). SCS may be enriched in
CO
2
fixation bacteria. However, there are no report
on the cbbL gene diversity from the SCS at the
moment.
In this study, the geographical distribution and
phylogenetic diversity of cbbL gene in sediments of
the South China Sea were determined with the aim
to broaden our view on the diversity of different
deep-sea habitat.
Su, J., Ming, H., Chen, Q., Jin, Y., Zhang, C., Guan, D. and Fan, J.
Distribution and Phylogenetic Diversity of CbbL Gene Encoding RuBisCo in the Deep-sea Sediments from the South China Sea.
In Proceedings of the International Workshop on Environment and Geoscience (IWEG 2018), pages 5-10
ISBN: 978-989-758-342-1
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
5
2 MATERIAL AND METHODS
2.1 Sample Collection
Deep-sea surface sediments (5 cm lower than the top
layer) were collected using a gravity box-coring
device from three sampling site in the Nansha area
of the South China Sea in the Spring of 2013.
Characteristics of three sampling sites were
described in Table 1.
Table 1: Sample information.
Sites Water
Depth(m)
pH eh
(mv)
Temperature Sample
describe
NSCA 1743 7.44 172 2.9 Yellow
mud
NSCE 1683 7.35 176 3.0 Brown
mud
NSCI 963 7.37 152 11.1 Red mud
2.2 DNA Extraction
Total genomic DNA was extracted from 5 g
sediments following with the Rapid Soil DNA
Isolation Kit (Sangon Biotech, Shanghai, China) and
further purified using the QIAquikPCR purification
kit according to the manufacturer’s directions.
2.3 Amplification, Cloning and
Sequencing of CbbL Gene
The purified DNA was used for cbbL gene
amplification using primers 595f and 1387r (Hung et
al, 2012). Thermo cycling reaction was as
followed:95℃ annealing for 5min;35 cycles of 94℃
for 30 s,52℃for 30 s,72℃for 1min;72℃for
10min.The PCR products was cloned using the
TOPO TA cloning kit (Invitrogen, Carlsbad, CA,
USA) and Escherichia coli TOP10 competent cells.
The inserted cbbL gene was sequenced by a
commercial company (Takara, Dalian).
2.4 Phylogenetic Analysis
Preliminary analysis of the sequences was
performed using BLAST (http://www.
ncbi.nlm.nih.gov/blast/). The nucleotide and inferred
amino acid sequences were aligned with sequences
from GenBank using CLUSTAL W (Thompson et
al., 1994). The aligned sequence with more than 97%
similarity was defined as one Operational
Taxonomic Unit (OTU). Phylogenetic tree of
bacteria harbouring cbbL gene was constructed
using neighbour-joining algorithm within the
MEGA 5.0 software. Alpha-diversity indices,
including Shannon, and Simpson values, were
subsequently calculated by Mothur.
3 RESULTS
3.1 Diversity Analysis of Cbbl Clone
Libraries
Bacterial cbbL sequences were obtained from all
three samples (NSCA, NSCE, NSCI). A total of 236
clones were achieved from three cbbL clone
libraries constructed from the SCS deep-sea
sediments. When analyzed using a 97% sequence
similarity cut off, the 236 total cbbL sequences
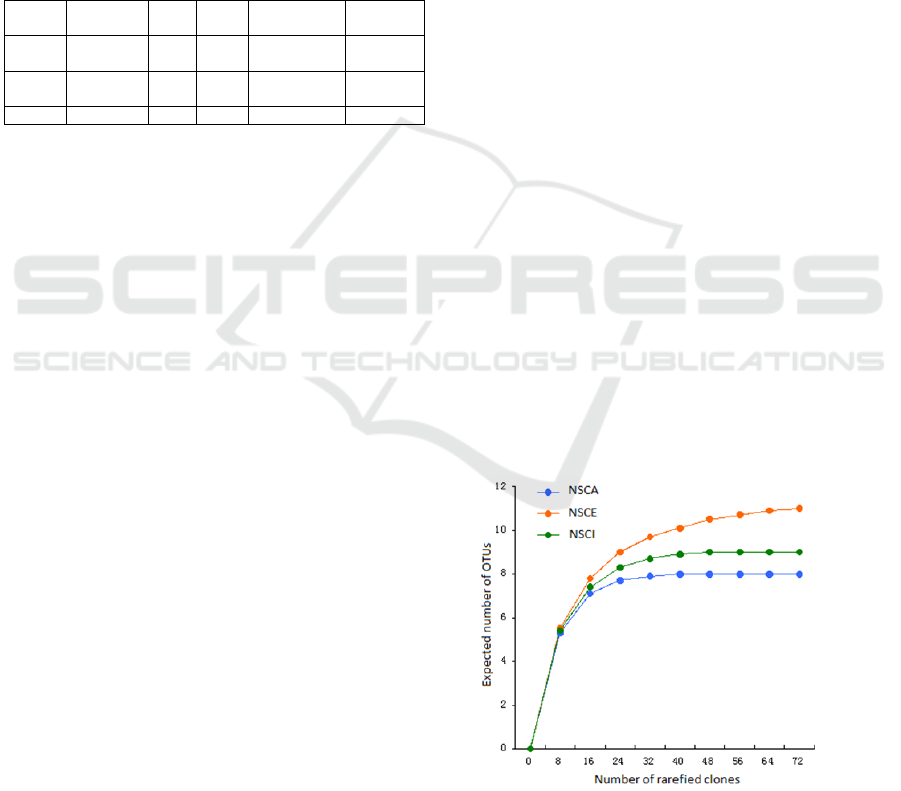
formed 28 OTUs. Rarefaction analysis of the clone
libraries showed that the species accumulation
curves were asymptotic for all three libraries (Figure
1), indicating a good coverage of the total cbbL gene
diversity in the SCS deep-sea sediments. The
estimated coverage values showed that more than
85% of bacterial cbbL diversity was captured (Table
2). This result indicates the capacity of cbbL clone
libraries is large enough for diversity analysis.
Shannon and Simpson indices were calculated to
evaluate the evenness and diversity of the cbbL at
each site. Shannon-Wiener index of cbbL clone
libraries from high to low is NSCE, NSCI and
NSCA respectively, suggesting the NSCE site
sediments is in the highest bacterial diversity among
the three sampling sites.
Figure 1: Rarefaction curves for the cbbL.
IWEG 2018 - International Workshop on Environment and Geoscience
6
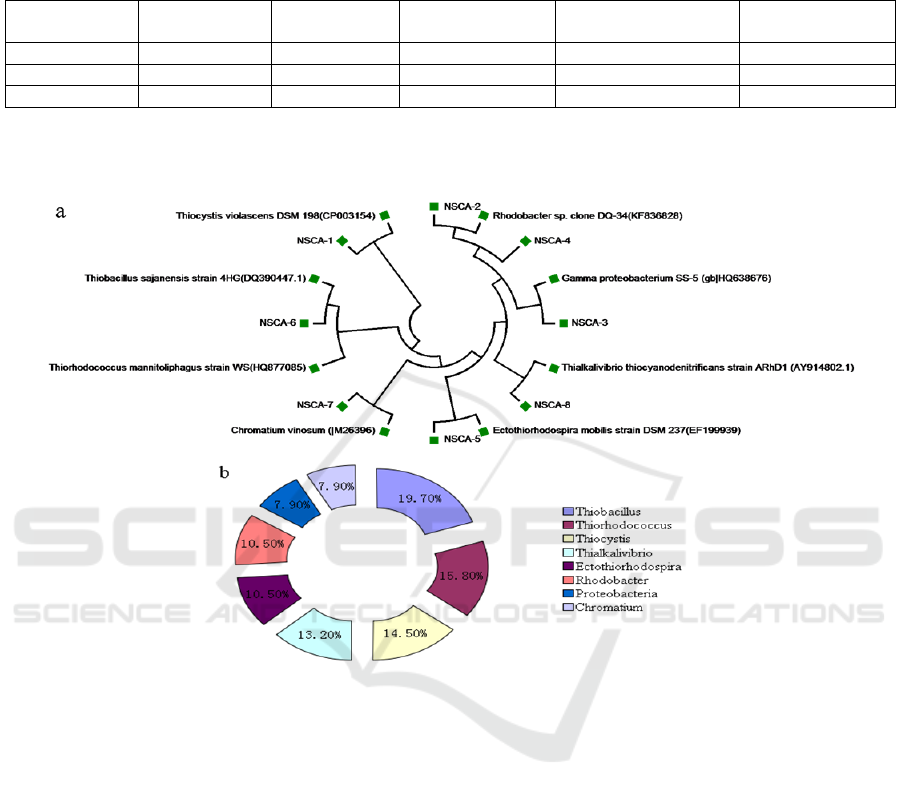
Table 2: Diversity indices for the cbbL clone libraries.
Clone library No. of clones No. of OTUs
a
Coverage (%)
b
Shannon-Wiener
index
c
Simpson index
NACS 76 8 89.4 2.03 0.86
NACE 80 11 86.2 2.2 0.88
NSCI 80 9 88.7 2.1 0.87
a
Percentage of coverage is the percentage of observed number of OTUs divided by the Chao1 estimated value
b
For the Shannon diversity index, a higher number represents greater diversity
c
For the Simpson diversity index, a higher number represents greater diversity
Figure 2: Phylogenetic analysis of bacteria harbouring cbbL gene derived from sediments of NSCA site.(a)
Phylogenetic tree based on the cbbL (translated amino acids) sequences;(b)Distribution of cbbL gene in deep-sea
sediments.
3.2 Phylogenetic Analysis of Cbbl
Genotypes
The expected size fragment was amplified with
DNA extracted from three sediment samples and
preliminary analysis of sequence yielded positive
results with the cbbL gene. A total of 28 OTUs from
three cbbL clone libraries covered 16 phylotypes:
Thiobacillus, Thiorhodococcus, Thiomonas,
Halothiobacillus, Halorhodospira halochloris,
Acidithiobacillus, Ectothiorhodospira, Thiobacillus
denitrificans, Nitrosomonas, Hydrogenophaga,
Thialkalivibrio denitrificans, Thiocys,
Thialkalivibrio, Thiobacillus denitrificans,
Chromatium and Rhodobacter. It may be concluded
that all of the cbbL sequences detected in Deep-sea
sediments belong to the Proteobacteria. The
phylogenetic analysis results were matching with the
above diversity indices, indicating the diversity of
the cbbL gene in the South China Sea.
The bacterial communities harbouring cbbL gene
derived from NSCA site were clustered into three
classes: α-Proteobacteria, β-Proteobacteria and γ-
Proteobacteria; 8 genera: Thiobacillus,
Thiorhodococcus, Thiocystis, Thialkalivibrio,
Ectothiorhodospira, Proteobacteria, Chromatium
and Rhodobacter (Figure 2). The predominant group
in NSCA site deep-sea sediments included
Thiobacillus, Thiorhodococcus, Thiocystis,
Thialkalivibrio, which was chemoautotrophic
bacteria involving in Calvin–Benson–Bassham cycle.
Distribution and Phylogenetic Diversity of CbbL Gene Encoding RuBisCo in the Deep-sea Sediments from the South China Sea
7
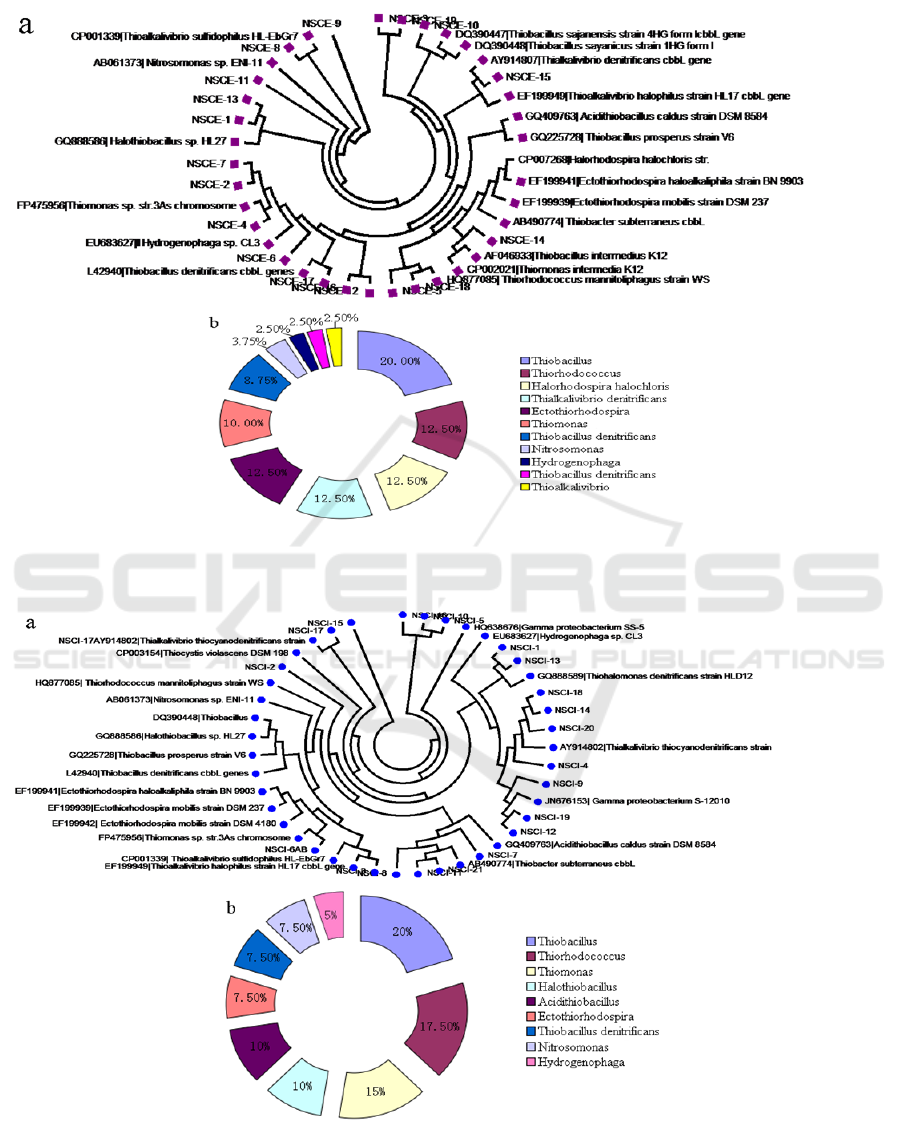
Figure 3: Phylogenetic analysis of bacteria harbouring cbbL gene derived from sediments of NSCE site.(a) Phylogenetic
tree based on the cbbL (translated amino acids) sequences;(b)Distribution of cbbL gene in deep-sea
sediments.
Figure 4: Phylogenetic analysis of bacteria harbouring cbbL gene derived from sediments of NSCI site.(a) Phylogenetic
tree based on the cbbL (translated amino acids) sequences;(b)Distribution of cbbL gene in deep-sea sediments.
IWEG 2018 - International Workshop on Environment and Geoscience
8

The bacterial communities harbouring cbbL gene
derived from NSCE site were clustered into two
classes:β-Proteobacteria and γ-Proteobacteria; 11
genera: Thiobacillus, Thiorhodococcus,
Halorhodospira halochloris, Thialkalivibrio
denitrificans, Ectothiorhodospira, Thiomonas,
Thiobacillus denitrificans, Nitrosomonas,
Hydrogenophaga, Thiobacillus denitrificans and
Thioalkalivibrio (Figure 3). The major bacterial
groups in the NSCE site sediments was found to
include Thiobacillus, Thiorhodococcus,
Halorhodospira halochloris, Thialkalivibrio
denitrificans, Ectothiorhodospira, also with low
abundant of Nitrosomonas, Hydrogenophaga,
Thiobacillus denitrificans and Thioalkalivibrio.
In the phylogenetic tree constructed from the
phylotypes of NSCE clone libraries, eleven OTUs
could be assigned to three classes: β-Proteobacteria,
γ-Proteobacteria and Acidithiobacillia; 9 genera:
Thiobacillus, Thiorhodococcus, Thiomonas,
Halothiobacillus, Acidithiobacillus,
Ectothiorhodospira, Thiobacillus denitrificans,
Nitrosomonas and Hydrogenophaga (Fig. 4).
Thiobacillus was the most dominant group and
accounted for 20% of in NSCI site deep-sea
sediments. Other predominant genera in NSCA site
deep-sea sediments included Thiorhodococcus and
Thiomonas, which were also chemoautotrophic
bacteria involving in Calvin–Benson–Bassham
cycle.
4 DISCUSSIONS
The RuBisCO gene were detectable in the SCS
deep-sea sediments and the general richness of the
cbbL gene was relatively high (from 0.11to 0.14
OTU per clone). Similar results were reported from
other habitats. The richness of cbbL genes Giri et al.
detected (0.12 OTU per clone) in Mono Lake was
comparable to the richness we observed, despite the
differences in habitat diversity (Giri et al., 2004).
Elsaied et al. identified the richness of cbbL genes
(0.10 OTU per clone) covering a range of habitats
associated with a hydrothermal vent site, including
sediment, overlying water, and as symbionts
(Elsaied and Naganuma, 2001). However, RuBisCO
genes with low richness were also observed in some
extreme habitats such as the deep-sea hydrothermal
vents, volcanic deposits and deep hypersaline anoxic
basin (Elsaied and Naganuma, 2001; Nanba et al,
2004; Elsaied et al, 2007; Wielen, 2006). Therefore,
the diversity of the cbbL gene may be correlated
with certain characteristics of the microbial habitats.
The amplicons of the cbbL gene all belonged to
form IA RuBisCO. This form is mainly found in
Alpha-, Beta- and Gammaproteobacteria, although a
few cyanobacterial sequences possess form IA as
well (Wielen, 2006). This study also indicated a
domination of the Proteobacteria distributed
throughout the SCS deep-sea sediments. Giri et al.
reported the similar results that the genus
Thiobacillus and Thiorhodococcus were the
dominant bacteria isolated from Mono Lake.
Thiobacillus-related RuBisCO were found to be
distributed globally and contribute to primary
production in the deep sea (Elsaied and Naganuma,
2001).Thiocystis with high proportion was detected
in NSCA site, while not detectable in NSCE and
NSCI. Rhodobacter as one genera of α-
proteobacteria was only present at the NSCA site,
also not detected in other two sites. The diversity of
bacterial populations in marine sediments maybe
due to the environmental characteristics difference
even in the same habitat. Among the detected groups
of the Gammaproteobacteria, the genera
Thioalkalivibrio were chemotrophic genus,
Halorhodospira and Ectothiorhodospira were
phototrophic genus.The 16 phylotypes that we
obtained from three SCS deep-sea sediments belong
to autotrophic bacteria and most were
chemoautotrophic bacteria. This was expected as
sampling site is located at 1000 m in the deep sea, a
depth at which light does not penetrate. Most of the
cbbL sequences detected in deep-sea sediments were
found belong to sulfur-oxidizing
Gammaproteobacteria and confirm the importance
of sulfur cycle bacteria in deep sea ecosystem.
Chromatium, Hydrogenophaga and
Ectothiorhodospira detected in this study were
facultative autotrophic bacteria. In conclusion, we
propose that the distribution of the deep-sea
RuBisCO genes cbbL may correlate with certain
characteristics of the microbial habitats.
ACKNOWLEDGEMENT
This work was supported by the National Key
Research Program (Grant 2016YFA0601400), the
State Oceanic Administration (Grant GASI-03-01-
02-05) of China and Key Laboratory for Ecological
Environmental in Coastal Areas (Grant 201813).
Distribution and Phylogenetic Diversity of CbbL Gene Encoding RuBisCo in the Deep-sea Sediments from the South China Sea
9

REFERENCES
Coffin M F 2004 Earth, oceans, and life: science of the
integrated ocean drilling program (IODP) Chikyu
Monthly 44 16
Dai X, Zhou H, Chen YQ 2002 A preliminary study on
16S rDNA diversity of bacteria in the Nansha marine
sediment, the South China Sea Progress in Natural
Science 12 479
Elsaied H E, Kimura H, Naganuma T 2007 Composition
of archaeal, bacterial, and eukaryal RuBisCO
genotypes in three Western Pacific arc hydrothermal
vent systems Extremophiles 11 191
Elsaied H, Naganuma T 2001 Phylogenetic diversity of
ribulose-1,5-bisphosphate carboxylase/oxygenase
large-subunit genes from deep-sea microorganisms
Applied and Environmental Microbiology 67 1751
Giri B J, Bano N, Hollibaugh J T 2004 Distribution of
RuBisCO genotypes along a redox gradient in Mono
Lake, California Applied and Environmental
Microbiology 70 3443
Hung W L, Wade W G, Chen Y, Kelly D P, Wood A P
2012 Design and evaluation of novel primers for the
detection of genes encoding diverse enzymes of
methylotrophy and autotrophy Polish journal of
microbiology 61 11
Kovaleva O L, Tourova T P, Muyzer G, Kolganova T V,
Sorokin D Y 2011 Diversity of RuBisCO and ATP
citrate lyase genes in soda lake sediments FEMS
microbiology ecology 75 37
Nanba K, King G M, Dunfield K 2004 Analysis of
Facultative Lithotroph Distribution and Diversity on
Volcanic Deposits by Use of the Large Subunit of
Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase
Applied & Environmental Microbiology 70 2245
Selesi D, Schmid M, Hartmann A 2005 Diversity of
Green-Like and Red-Like Ribulose-1,5-Bisphosphate
Carboxylase/Oxygenase Large-Subunit Genes (cbbL)
in Differently Managed Agricultural Soils Applied and
Environmental Microbiology 71 175
Shively J M, Devore W, Stratford L, Porter L, Medlin L,
Jr S E S 1986 Molecular evolution of the large subunit
of ribulose-1, 5-bisphosphate carboxylase/oxygenase
(RuBisCO) FEMS microbiology letters 37 251
Wielen P W van der 2006 Diversity of ribulose-1,5-
bisphosphate carboxylase/oxygenase large-subunit
genes in the MgCl2-dominated deep hypersaline
anoxic basin discovery FEMS microbiology letters 259
326
Yuan H, Ge T, Chen C, O'Donnell A G, Wu J 2012
Significant role for microbial autotrophy in the
sequestration of soil carbon Applied and
Environmental Microbiology 78 2328
IWEG 2018 - International Workshop on Environment and Geoscience
10
