
A Semantic-based Approach for Facilitating Arbovirus Data Usage
Aparecida Santiago
1
, André Alencar
1
, Amanda Souza
2
, Erika Araruna
2
, Isabel Fernandes
2
and Damires Souza
1
1
Academic Unit of Informatics, Federal Institute of Paraiba, João Pessoa, Brazil
2
Medical Science Center, Federal University of Paraíba, João Pessoa, Brazil
damires@ifpb.edu.br
Keywords: Semantics, RDF Data, Data Usage, Metadata Reuse, Arbovirus Data.
Abstract: Today’s continuous growth for healthcare information entails an increasing need for using large amounts of
data. Particularly, the incidence of arboviruses has been on the rise in some countries, what causes specific
needs for studies and definitions of public strategies. In this light, providing a computational platform for
usage and reuse on arboviruses related data may help matters. The idea is that different applications and
users can make use of that data in diverse ways. In this work, we propose a semantic-based approach for
facilitating use and reuse of arboviruses related data. We present the definitions underlying our approach,
examples illustrating how it works, and some promising results we have obtained.
1 INTRODUCTION
Today’s continuous growth for healthcare
information entails an increasing need for using
large amounts of data, which may come from
different data sources. Particularly, the data scenario
associated with vector-borne diseases is currently
causing specific needs for studies and definitions of
public strategies in some countries.
Vectors are living organisms that can transmit
infectious diseases between humans or from animals
to humans. Mosquitoes are examples of vectors. The
Aedes aegypti mosquito is a vector, which transmits
four different diseases commonly called as
arboviruses (Fletcher, 2017), as follows: Dengue
fever, Yellow fever, Zika virus and Chikungunya
fever. Given the spread of these diseases in some
countries, it is necessary to improve control
strategies. Providing data sharing and usage, by
means of a computational platform, may enhance the
development of applications and data analytics in
such a way that healthcare managers and doctors
may take important decisions.
A large amount of information on arboviruses is
available on the Web in sources such as sites. Data
are also found in specific databases, many of them
local to some hospitals. With this diversity of data
sources, with their own terminological definitions, it
is hard for a computational application to solve the
conflicts arising from the existing heterogeneities
and achieve a common understanding of the data
(Bansal and Kagemann, 2015).
To help matters, it is necessary to collect and
integrate existing data on arboviruses and share them
in a way that makes them feasible for easier usage.
In order to make the computational effort smaller in
the development of a solution, some principles and
technologies derived from the Semantic Web (Heath
and Bizer, 2011) can be employed. At first, the data
should be described semantically, i.e., according to a
common understanding, what facilitates their
processing and reuse (Bansal and Kagemann, 2015).
To this end, it is necessary to choose and employ a
domain vocabulary in order to provide semantic
reference to the data. These data are usually
converted to the RDF data model in order to enable
the semantic description (Lóscio et al., 2017).
With this scenario in mind, we define two main
research problems that have guided our work, as
follows: (i) How to provide a standard vocabulary
on arboviruses so that researchers, doctors,
healthcare managers as well as software agents can
use it as a reference for data conversion and
sharing? And (ii) Given arbovirus related data,
semantically described in RDF, can they be used in
order to facilitate the development of useful
applications on diseases control?
Santiago, A., Alencar, A., Souza, A., Araruna, E., Fernandes, I. and Souza, D.
A Semantic-based Approach for Facilitating Arbovirus Data Usage.
DOI: 10.5220/0006799706710678
In Proceedings of the 20th International Conference on Enterprise Information Systems (ICEIS 2018), pages 671-678
ISBN: 978-989-758-298-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
671

In this work, we present a semantic-based approach
that aims to facilitate usage and reuse of arboviruses
related data and metadata. Semantic technologies are
employed for modelling relevant information by
means of an ontology, which implements the domain
vocabulary. The approach includes a tool, which is
able to convert CSV data into RDF. In order to
verify the usefulness of the converted data, a web
application, which provides arbovirus information
visualization, has also been developed and
evaluated. In addition, some experiments have been
accomplished.
Our contributions are summarized as follows: (i)
we introduce the ARBO ontology; (ii) we propose a
semantic-based approach to convert arbovirus data
into RDF ones; (iii) we present an application, which
provides useful information based on the produced
RDF data; and (iv) we describe accomplished
evaluations w.r.t. the proposed approach.
The remainder of this paper is organized as
follows: Section 2 introduces some background
concepts, a motivating scenario and related work;
Section 3 presents the proposed approach; Section 4
shows some obtained results and describes the
accomplished evaluations; Finally, Section 5 draws
our conclusions and points out some future work.
2 CONCEPTS, SCENARIO AND
RELATED WORK
In this section, we provide some concepts and
recommended practices for sharing data on the Web.
We also provide a motivating scenario and discuss
some related works.
2.1 Data on the Web
The Web has evolved into an interactive information
network, allowing users and applications to share
data on a massive scale. To help matters, the
Semantic Web and the Linked Data principles define
a set of practices for publishing structured data on
the Web aiming to provide an interoperable Web of
Data (Heath and Bizer, 2011). These principles are
based on technologies such as HTTP, URI and the
RDF data model. By using the RDF model, data or
resources are published on the Web in the form of
triples (composed by a subject, a predicate and an
object). Each resource is identified by means of an
URI. In order to achieve this, it is necessary to
convert data, which are originally in other format
(e.g., CSV), to RDF data.
In order to make data available and feasible for
reuse, another semantic web principle is to organize
data in such a way that they can be interpreted and
used meaningfully without human intervention
(Bansal and Kagemann, 2015). This is achieved by
adding data about data, i.e., by adding metadata to
refer semantically the data.
To clarify matters, the World Wide Web
Consortium (W3C) defines some best practices to
facilitate sharing data on the Web (Lóscio et al.,
2017). These best practices cover diverse aspects
related to data publishing and consumption, like data
formats, data access, data identification and
metadata provisioning. One of the recommendations
regards the use of open domain vocabularies in order
to semantically refer the data, when data are
converted to RDF. To this end, it is essential to take
into account the knowledge domain (e.g., “Health”,
“Music”) in which the data exist and choose the
appropriate domain vocabularies. Vocabularies are
usually developed as ontologies, which represent a
formal, explicit specification of a conceptualization
(Gruber, 2009). An ontology provides definitions of
terms in a given data domain as well as the
relationships that link these terms to each other.
Other W3C recommendation regards facilitating
data consumption. In this sense, it is important to
make data available through APIs (Application
Programming Interfaces), developed for such
purpose, especially if data are large, frequently
updated, or highly complex.
2.2 Motivating Scenario
Collecting and integrating data on diseases, such as
arboviruses, become relevant to some specific
applications, particularly in times of their high
incidence in some countries. We have observed the
need of data analytics on these diseases not only by
healthcare agency managers but also by healthcare
professionals. They have to plan and study
preventive measures in order to fight diseases
occurrences and consequences.
Some data on arboviruses are already published
on the Web as open data. Nevertheless, in some
governmental states as ours, there are no open data
portals with such data. In this work, we have
obtained data directly from the state healthcare
agency. As an illustration, excerpts from the
obtained data are depicted in Figure 1.
Lines in Figure 1 represent patients and
occurrences of disease notification (dengue or
chikungunya). For each patient, symptoms (most
columns) are set according to medical anamnesis.
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
672

Existing symptoms are included as “1” value; on the
other hand, if a given symptom is not present in
patient complains, it is defined as “2” value. To
facilitate understanding, we present the english
meaning of the symptom terms present in the data
sets (properly ordered), as follows: fever (febre),
myalgia (mialgia), headache (dor de cabeça),
exanthema (exantema), vomit (vômito), arthritis
(artrite), arthralgia (artralgia), petechiae (petequia),
leukopenia (leucopenia) and tie proof (prova do
laço).
Figure 1: Excerpts from Real Arbovirus Data.
In order to have an integrated view of the data from
the data sources, it is necessary to deal with phases
such as data Extraction, Transformation and Load
(ETL) (Bansal and Kagemann, 2015). Each phase
has specific technical issues to be addressed. To
facilitate this process, identifying the relevant data to
extract, creating feature extractors and converters,
and building a domain vocabulary to align the data
are usual steps to be done.
We use the presented data scenario for
motivating this work and also for demonstrating how
the proposed approach works in a real-world data
environment. Nevertheless, the proposed approach
may be instantiated in any arbovirus data scenario.
2.3 Related Work
Literature about disease ontologies is not new and
some works already provided useful artifacts. In this
section, we briefly resume some relevant work in
this data domain. We also discuss works regarding
data conversion, semantic platforms for health and
use of data on applications and analytics.
Some studies have been carried out on the
creation of health ontologies, such as the IDODEN
(ontology for Dengue) and the IDO (Infectious
Disease Ontology) regarding viruses in general
(Bioportal, 2017). In addition, other ontologies
related to this knowledge domain are IDOMAL, an
ontology on malaria information, which extends
IDO, and the MEDDRA, MESH and SNMI, which
are ontologies that provide a terminology for
cataloging medical information (Bioportal, 2017).
The ontologies most related to our work are the
IDO, IDODEN and DOID. IDO is a consortium of
infectious disease ontologies, among which are
currently being developed ontologies for Dengue
and Malaria. IDODEN is an ontology for Dengue,
addressing clinical aspects, which extends the IDO
ontology. The DOID ontology is defined by
following a scope of human diseases in general.
With respect to general computational
approaches, Dragoni et al. (2017) developed an
architecture for supporting the monitoring of people
and for persuading them to follow healthy lifestyles.
To this end, they used semantic technologies for
modeling relevant information and for fostering
reasoning activities. Chun and MacKellar (2012)
developed a system, which integrates information
from some sites such as PatientsLikeMe (Patients,
2018) and PubMed (Pubmed, 2018). It can be used
to annotate a variety of text based blogs.
Regarding tools developed to provide data
conversion to RDF, the LinkMapia application
(Sacenti and Fileto, 2014) is an example. It converts
geographic data into linked data. It also filters data
to align them with existing collections of linked
data. Regarding applications which consume data on
diseases, two examples are provided. Kaieski
developed the Vis-Health application, an open
source system in which records from public health
are used to provide some analyses (Kaieski, 2014).
Varela (2016) implemented an application for
tracking and presenting data on arboviruses, with the
purpose of informing, by using maps and charts the
list of hospitals that received infected patients.
These last two applications do not deal with RDF
data, differently from ours. Also they produce
distinct kinds of data analyses. Our approach uses a
specific ontology to assist real data to be converted
to integrated RDF data as part of the transformation
step of an ETL process. Based on the produced RDF
data, an information visualization application has
been developed as a means to validate data
consuptiom and usage.
3 PROPOSED APPROACH
Vocabularies provide the semantic glue enabling
data to become more meaningful data. With the
emergence of open vocabulary repositories, many
vocabularies are being published and similar ones
are being grouped together usually on the web.
Examples of such repositories are the Linked Open
A Semantic-based Approach for Facilitating Arbovirus Data Usage
673

Vocabulary (LOV, 2017) and, more specifically, the
Bioportal, which is related with the health
knowledge domain (Bioportal, 2017).
As a result, finding a suitable vocabulary for
publishing a specific dataset in RDF has become
easier, although it is usually necessary to select one
with a wide consensus in the community. However,
in case of unavailability of a suitable one, or when it
does not cover completely a given set of knowledge
domain terms, it is necessary to build a new one and
reuse terms which already have been defined.
Although there are some specific vocabularies
regarding arboviruses such as dengue, to the best of
our knowledge, we could not find specific ones
related to the recent arboviruses, i.e., to chikungunya
fever and zika virus. As a result, we have worked on
an ontology, which covers the domain of arboviruses
in terms of general kinds of diseases, symptoms,
signals, exams, severity of the diseases and other
additional information. All developed ontology
terms were suggested by medical specialists. In the
following, we present the ARBO ontology. Then we
describe how the ontology is used along with the
data conversion and publication process.
3.1 The ARBO Ontology
Based on some methodologies of ontology
engineering (Sure et al., 2009), we have instantiated
an iterative and incremental process to develop the
ARBO ontology. The ontology building process
includes the following steps:
I. Determination of the knowledge domain
and its scope: in this work, the knowledge domain
refers to the viruses group named as arboviruses.
II. Enumeration and definition of important
terms w.r.t. concepts and properties of the domain at
hand (conceptual domain model).
III. Survey of existing and relevant
vocabularies for allowing the reuse of some terms.
IV. Definition of classes, hierarchies and
properties. A mapping between the candidate terms
and the terms identified in the domain conceptual
model was performed.
V. Validation of the ontology terms by
domain experts. In our case, healthcare doctors and
researchers have provided such validation.
The ARBO ontology makes reuse of terms from
existing ontologies such as IDOMAL, IDODEN and
DOID instead of creating duplicates of terms. Thus,
it ensures interoperability with already existing
infectious disease ontologies. Table 1 presents the
list of reused ontologies and the number of terms
reused from each one. Additionally, it shows the
number of specific domain terms (155), which have
been originally created in the ARBO ontology. By
adding all the terms reused and created, the ARBO
ontology is now composed by 218 terms, of which
63 are reused and 155 were newly created for this
ontology. The main concepts of the ARBO ontology
are depicted in Figure 2, according to the ontograf
notation (Ontograf, 2017).
Table 1: Vocabularies and the Number of Reused or
Created Terms.
Ontology
Number
of Terms
DOID 15
IDODEN 8
IDOMAL 3
SYMP 13
MESH 5
MEDDRA 17
SNMI 1
SNOMEDCT 1
ARBO 155
The ARBO vocabulary comprises some primary
concepts such as Disease, Disease by Infectious
Agent, Viral Infectious Disease, Arboviruses, Pre-
existing diseases, Patient, Symptom, Signal,
Chikungunya, Zika, Dengue and Yellow Fever.
Properties and relationships are defined by means of
data and object properties, respectively.
We have identified these terms with the
assistance of two medical experts in virus caused
diseases. In accordance with their guidance, we have
also differentiated symptoms from signs or signals.
The signs and symptoms described in this work
correspond to the observations from the doctor and
the complaints presented from the patient in the
medical appointment, respectively. The signs refer
to more objective and direct data, which can be
noticed by the doctor, nurses and relatives, along the
physical exam or at home. The symptoms, in
contrast, are subjective and only can be described by
the patient, as the characteristics or manifestations of
the disease in his/her body.
At the ARBO documentation (ARBO, 2017), we
depict terms that have been defined not only to
symptoms but also to signs. Both are very important
since they provide means to doctors to understand
patient complains. The ARBO ontology will be
published in the main open vocabulary repositories
such as LOV and Bioportal. Descriptive metadata
have been included in the ARBO ontology in order
to provide information such as the ontology creators,
publisher, version number, and date of publication.
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
674
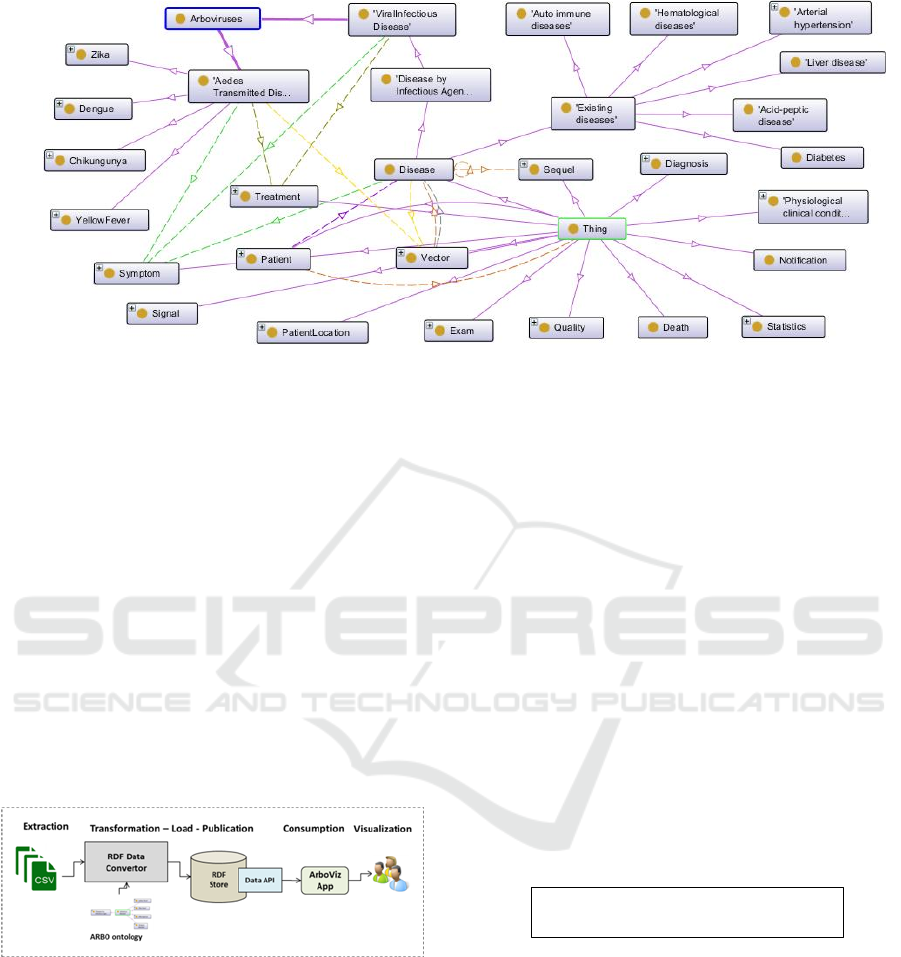
Figure 2. Main Concepts of the ARBO Ontology.
3.2 The Data Publication Process
The main idea underlying our approach is to bring
the knowledge domain semantics into the data ETL
process aiming to facilitate data publication and
consumption. The activity of converting different
data sources on arboviruses produces an integrated
view of the data defined in terms of a given domain
vocabulary. In this work, we use the ARBO
ontology to provide that means.
The proposed semantic data publication process
consists of the three ETL major phases along with
the data publication and consumption phases, as
depicted in Figure 3. The use of semantic
technologies is introduced in the Transformation
phase as a means to enhance data conversion. Phases
are discussed in the following.
Figure 3: The Data Publication and Consumption Process.
Data Extraction
In the data extraction step, instance data along with
their properties (metadata) are extracted from
existing datasets. In this work, CSV datasets are
considered. Data cleaning tasks are applied in order
to prepare the data. Metadata are identified
according to the names of the properties (columns),
which compose the CSV file. A developed extractor
provides the selection of the properties and data to
be converted.
Data Transformation
In this step, the extracted metadata along with their
corresponding data are converted to RDF triples. At
first, the matching of extracted CSV metadata
against the ARBO vocabulary terms is done. To
assist this process, a user known as Domain Expert
(DE) is needed. The DE has an understanding of the
content to be converted and the knowledge domain
underlying the data. The DE assists the matching
activity by pointing out the correspondences
between the extracted metadata and the ontology
properties. The output of the matching process is
called an alignment. It contains a set of equivalence
correspondences indicating which properties
correspond to each other. Examples of these
correspondences are shown in Figure 4. This
alignment is saved and used later. Then, based on
the identified correspondences, for each property
(e.g., symptoms, signs) and, for each row (e.g.,
patients), RDF triples are generated.
Figure 4. Examples of correspondences between original
data properties and the ARBO terms.
Data Load and Publication
The generated RDF dataset is persisted in an RDF
store and made available on the web as linked data.
This means that it is available for querying via a
SPARQL endpoint.
Data Consumption and Visualization
The RDF dataset on arboviruses has a SPARQL
endpoint that allows its consumption. Developers
ID_MUNICIP arbo:hasCity
DT_NOTIFIC arbo:dataOfNotification
A Semantic-based Approach for Facilitating Arbovirus Data Usage
675

have programmatic access to the data on arboviruses
for use in their own applications. As an initial
example of a consumption application, the arboviz
application (described in Section 4.1), has been
developed.
4 RESULTS AND EVALUATION
In this section, we describe some implementation
and evaluation results.
4.1 Developed Tools
We have developed the data conversion process
within a tool implemented in PHP. In this version, it
is able to convert CSV files to the RDF model, using
information from the knowledge domain of the data.
Although in this work we have used the ARBO
ontology and datasets provided by the state agency,
the tool is able to receive as input any CSV file
along with the domain ontology to be considered
(any) in order to provide the data conversion. Thus,
it may be used in any data domain.
Regarding the scenario illustrated in Figure 1
(part of the provided datasets), the tool is able to
generate the RDF dataset for each one of the CSV
files. The datasets refer to the years 2015, 2016 and
2017 and to the disease notifications for Dengue and
Chikungunya fever.
As an illustration, we provide an excerpt from an
RDF dataset with respect to the occurrences of
Dengue in 2015 (Figure 5). The dataset is serialized
in RDF/Turtle. In this example, there is one patient
with related data. The patient refers to the resource
idoden:IDOMAL_0000603#0 (subject), which has
two predicates and their respective objects:
arbo:has_city
http://siderg.com.br/arbo/ID_MN_RESI/LASTRO,
and doid:has_symptom
http://siderg.com.br/arbo/CEFALEIA.
We have implemented a data consumption
application named as arboviz (arboviz, 2017). It was
developed in PHP, and it consumes data from the
RDF dataset with information about arboviruses
occurrences in the state of Paraíba, Brazil. It uses a
SPARQL endpoint to this end.
The main goal of arboviz is to provide easy and
accessible information visualization, with
explanatory texts and analytics generated from the
data present in the produced RDF dataset. One of the
produced views is depicted in Figure 6. It depicts the
main symptoms, regarding dengue and chikungunya
Figure 5. Excerpt from a generated RDF dataset.
diseases. To this end, it uses a word cloud, which is
built by considering the most cited symptoms in the
RDF dataset. Each symptom is printed in a given
font and scaled by a factor roughly proportional to
its number of occurrences in the underlying dataset.
Regarding dengue disease, the most referred
symptoms are (properly ordered) the following:
fever, headache, myalgia, arthralgia, arthritis,
nausea, back pain and vomit. Regarding
chikungunya, the most cited symptoms are (properly
ordered) the following: fever, myalgia, headache,
arthralgia, back pain, nausea, arthritis, and vomit.
Figure 6. Example of a Data View in the Arboviz App.
4.2 Evaluation
We have conducted two main types of evaluation to
verify the effectiveness of the proposed approach
and applications. The former regards evaluating if
the ARBO ontology indeed makes difference when
converting the data. The latter is concerned with the
usefulness of the arboviz application and,
consequently, the produced RDF data.
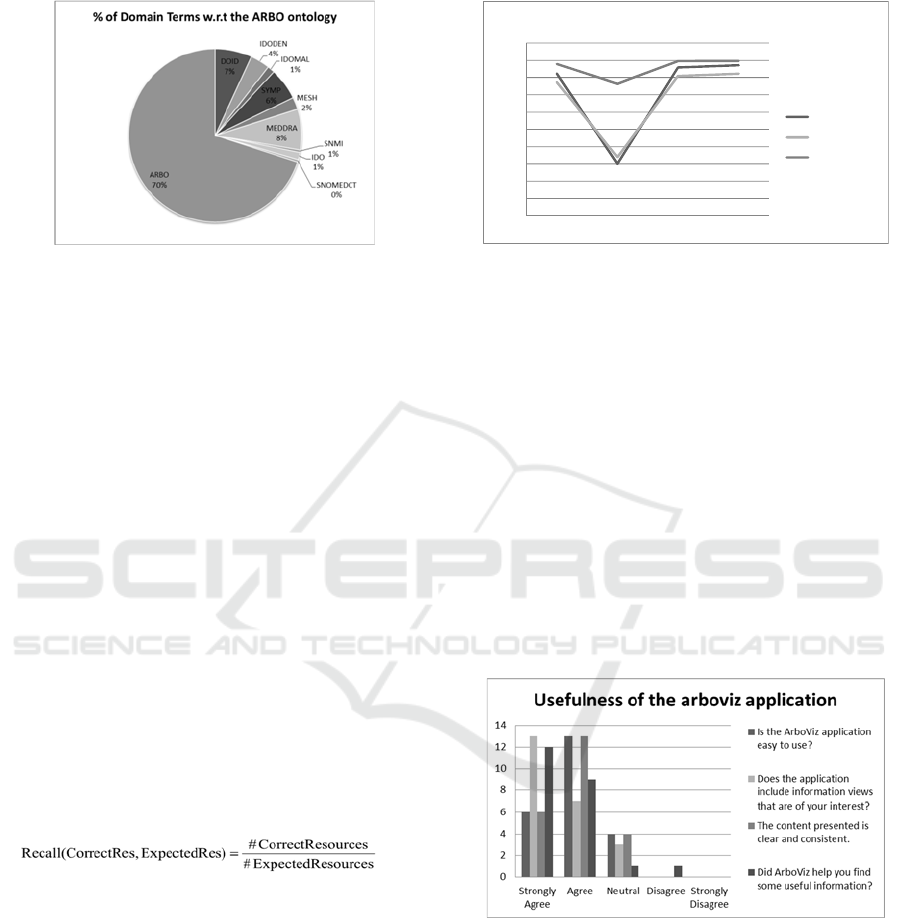
To verify the former claim, two experiments
were done. Firstly, we performed a comparison
among the domain terms which have been used to
compose the ARBO ontology (Figure 7), shown in
Table 1. In addition to the number of terms depicted
in Table 1, we have also considered the number of
common terms between the IDO and the ARBO
ontology, what results in 03 terms. We observe that
some domain terms are rather important to the
composition of the final one (e.g.,
doid:hasSymptom), although most terms (70%) had
to be indeed created in the ARBO ontology, in
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
676
accordance with recommendations provided by
healthcare experts.
Figure 7. Number of Used Terms w.r.t the ARBO
Ontology Composition.
We have also conducted an experiment to verify the
degree of recall regarding the data conversion when
considering some ontologies versus the ARBO one.
In this particular evaluation, we used four CSV files
regarding arboviruses notifications. The first one is
identified as dengue2015 and contains 30.359 rows.
The second one is identified as dengue2016 and
contains 45.114 rows. The third dataset is named as
chikungunya2016 and is composed by 14.026 rows.
The last dataset is called chikungunya2017 and holds
884 rows. As domain vocabularies to be verified, we
have used the IDODEN, IDO and ARBO.
We consider recall as the ratio of correctly found
resources (true positives) over the total number of
expected resources (true positives and true
negatives) (Rijsbergen, 1979). To achieve the
expected number of resources, we have produced
gold standards regarding the RDF data generation
for each evaluated vocabulary versus dataset to be
converted. These gold standards have been manually
produced by participants of our research group. The
recall formula is presented in the following.
Where
#CorrectResources is the number of correct returned
resources (URIs);
#ExpectedResources is the total number of all possible
resources (URIs) that could be generated.
A summary of the results regarding the recall
measure along with the usage of IDODEN, IDO and
ARBO vocabulary for the four datasets is shown in
Figure 8. We are able to observe that the usage of a
suitable domain vocabulary makes all the difference.
In this work, we have defined a specific vocabulary
by making reuse of recommended terms when
possible. New terms which belong to health data
sources have been defined in the ARBO Vocabulary,
based on expert advice. As a result, it has covered
almost 100% of the required data.
Figure 8. Recall w.r.t. the choice of a domain vocabulary
on arboviruses.
In addition, we have invited some users to evaluate
the arboviz application w.r.t. its usefulness. The user
group was composed by a total of 24 persons by
means of general users (56%), healthcare managers
(22%) and healthcare professionals (22%). To
perform the evaluation, they used the arboviz
application and then they filled out a questionnaire.
They filled out a questionnaire stating their
opinions on the interface usability, the provided
information views and also on the ontology terms (in
case of healthcare professionals). They were also
asked to provide comments pointing out what they
most liked or not, along with suggestions. Among
the presented questions, four main ones are depicted
in Figure 9 with their respective answers.
Figure 9. Evaluation of the arboviz application w.r.t. its
usefulness.
In terms of easiness of use, information views and
content presentation, the users provided a great
impression. Particularly in terms of whether there is
useful information, only two persons considered the
application not providing that feature. In case of
healthcare professionals, they were also asked if the
ARBO vocabulary covers the major terms in the
area. All of them confirmed that the ontology is
complete w.r.t. the arboviruses domain area.
0,820
0,840
0,860
0,880
0,900
0,920
0,940
0,960
0,980
1,000
1,020
1234
Recallw.r.t.thechoiceofadomainvocabulary
RecallIDODEN
RecallIDO
RecallARBO
A Semantic-based Approach for Facilitating Arbovirus Data Usage
677

Regarding what the users most liked, they said:” the
application is simple and easy of finding
information; the symptoms chart, which shows the
symptoms associated with the body parts, is really
interesting, the presented content is useful and
integrates information from different sources in
unified views”. The biggest complaints were
regarding the “lack of responsiveness of the
application interface”.
5 CONCLUSIONS AND
FURTHER WORK
The publication and consumption of data on
arboviruses is indeed an important issue. Based on a
built domain ontology, we have developed an ETL
data process, which provides publication and
consumption of arbovirus related data and,
consequently, facilitates their usage. Data published
in RDF have potential to be used in many ways and
to facilitate the creation of data-driven applications
such as the described arboviz application.
Accomplished experiments show that our
approach is promising. By using the proposed
domain ontology, it is able to produce a recall
measure regarding data conversion of almost 100%
w.r.t. the source data. In addition, an evaluation
carried out with real users showed that the arboviz
application is useful since it provides kinds of
information views that unify data by presenting them
in different perspectives.
For future work, we intend to convert more real
data on arboviruses and deal with other data
analytics on them. In addition, the ARBO ontology
will be exposed as a web service.
REFERENCES
ARBO Documentation, 2017. Available at
http://siderg.com.br/arboviz/ontology/views.php. Last
access on December, 2017.
Arboviz, 2017. Available at http://siderg.com.br/
arboviz/index.php. Last access on December, 2017.
Bansal, S., Kagemann, S., 2015. Semantic Extract-
Transform-Load framework for Big Data Integration.
In Computer 48(3):42-50.
Bioportal, 2017. Available at https://bioportal.
bioontology.org/. Last access on December, 2017.
Chun, S., MacKellar, B., 2012. Social Health Data
Integration using Semantic Web. In Proceedings of the
27th Annual ACM Symposium on Applied Computing.
SAC.
Dragoni, M., Bailoni, T., Eccher, C.,Guerini, M.,
Maimone, R., 2017. A Semantic-enabled Platform For
Supporting Healthy Lifestyle. In Proceedings of the
ACM Symposium on Applied Computing. SAC.
Fletcher, J., 2017. Arboviruses: Types, symptoms, and
transmission. Available at
https://www.medicalnewstoday.com/articles/318645.p
hp. Last access on December, 2017.
Gruber, T., 2009. Ontology. Encyclopedia of Database
Systems, pp 1963-1965.
Heath, T., Bizer, C., 2011. Linked Data: Evolving the Web
into a Global Data Space, Morgan & Claypool
Publishers. 1
st
edition.
Kaieski, N., 2014. Vis-Saúde - Uma metodologia para
visualização e análise de dados de saúde pública.
Master Thesis, UNISINOS.
Lóscio, B., Burle, C., Calegari, N., 2017. W3C Data on
the web best practices. Available at:
https://www.w3.org/TR/dwbp/. Last access on
December, 2017.
LOV, 2017. Linked Open Vocabularies. Available at:
https://lov.okfn.org/dataset/lov/. Last access on
December, 2017.
Ontograf, 2018. The Ontograf plugin. Available at
https://protegewiki.stanford.edu/wiki/OntoGraf. Last
access on January, 2018.
Patients, 2018. Available at https://patientslikeme.com/.
Last access on January, 2018.
Pubmed, 2018. Available at
https://www.ncbi.nlm.nih.gov/pubmed/. Last access
on January, 2018.
Rijsbergen, C., J., 1979. Information Retrieval, MA:
Butterworths. Stoneham, 2
nd
edition.
Sacenti, J., Fileto, R., 2014. LinkMapia: Uma Abordagem
para Converter Dados Geográficos Livremente
Anotados em Dados Ligados. In: X Escola Regional
de Banco de Dados, 2014.
Sure, York., Staab, Steffen., Studer, Rudi., 2009. Ontology
Engineering Methodology. In Staab, Steffen & Studer,
Rudi (eds.) Handbook on Ontologies. Springer-Verlag,
Heidelberg, 2
nd
edition. ISBN 978-3-540-70999-2.
Varela,V., 2016. Rastreamento endêmico da dengue, Zika
e chikungunya via Android e sistema de informação
geográfica (SIG). Undergraduate Work. UNB.
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
678
