
VarSearch: Annotating Variations using an e-Genomics Framework
José Fabián Reyes Román
1,2
, David Roldán Martínez
1
, Alberto García Simón
1
, Urko Rueda
3
and Óscar Pastor
1
1
Research Center on Software Production Methods (PROS), Universitat Politècnica de València, Valencia, Spain
2
Department of Engineering Sciences, Universidad Central del Este (UCE), San Pedro de Macorís, Dominican Republic
3
Department of Information Systems and Computation (DSIC), Research Center on Software Production Methods (PROS),
Spain, Valencia
Keywords: e-Genomics, EGF, GeIS, Variation, Conceptual Modeling, CMHG, Precision Medicine.
Abstract: Nowadays experts in the genomics field work with bioinformatics tools (software) to generate genomic
diagnoses, but the reality is that these solutions do not fully meet their needs. From the perspective of
Information Systems (IS), the real problems lie in the lack of an approach (i.e., Software Engineering
techniques) that can generate correct structures for data management. Due to the problems of dispersion,
heterogeneity and the inconsistency of the data, understanding the genomic domain is a huge challenge. To
demonstrate the advantages of Conceptual Modeling (CM) in complex domains -such as genomics- we
propose “VarSearch”, a web-based tool for genomic diagnosis that incorporates the Conceptual Model of
the Human Genome (CMHG) and takes advantage of Next-Generation Sequencing (NGS) for ensuring
genomic diagnostics that help to maximize the Precision Medicine (PM).
1 INTRODUCTION
The study and understanding of the human genome
could probably be considered one of the great
challenges of our century. Thanks to the advances in
NGS (Next Generation Sequencing) (Mardis, 2008),
there has been considerable growth in the generation
of genomic and molecular information. In addition,
the interactions that are available with this genomic
knowledge have a direct impact on the medical
environment and Precision Medicine (PM) (Grosso,
2016).
The application of Conceptual Modeling (CM)
(Olivé, 2007) techniques to the genomic domain
now provides solutions and optimizes some of the
processes carried out by experts (i.e., in genetic
laboratories and hospitals), and helps to solve the
problems that arise in handling the large amounts of
information from different sequencing methods. The
use of advanced Information System (IS)
engineering approaches can be useful in this domain
due to the huge amount of biological information to
be captured, understood and effectively managed. A
considerable part of modern Bioinformatics is
devoted to the management of genomic data. The
existence of a large set of diverse data sources
containing large amounts of data in continuous
evolution makes it difficult to find convincing
solutions (Reyes Román et. al., 2016). When we
addressed this problem from the IS perspective, we
understood that precise CMs were required to
understand the relevant information in the domain
and to clearly fix and represent it to obtain an
effective data management strategy.
Research and genetic diagnoses are typical
examples of the work done by experts -biologists,
researchers or geneticists- every day. However,
some information is required to perform these tasks.
Where are these data? Currently, this information is
dispersed in genomic repositories including web
sites, databanks, public files, etcetera, which are
completely heterogeneous, redundant, and
inconsistent (containing partial information). In
addition, most of these just focus on storing specific
information to solve a specific problem (e.g. YHRD,
designed to store Y chromosome haplotypes from
global populations, https://yhrd.org/). Due to these
characteristics, we are able to estimate the difficulty
of experts in finding and manipulating certain
genomic information, making this goal almost
impossible to achieve. Another relevant factor in the
domain is the constant growth and updating of the
328
Reyes Román, J., Roldán Martínez, D., García Simón, A., Rueda, U. and Pastor, Ó.
VarSearch: Annotating Variations using an e-Genomics Framework.
DOI: 10.5220/0006781103280334
In Proceedings of the 13th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE 2018), pages 328-334
ISBN: 978-989-758-300-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
data (i.e., biological concepts). The use of standard
definitions of concepts is not mandatory, so that
sometimes the same term can have different
definitions, in which case the meaning of the
concept depends on the interpretation given to it by
the expert. After studying this situation, we decided
to develop a Genomic Information System (GeIS) in
the form of the VarSearch tool (Reyes Román et. al.,
2018) for data treatment and management. This
system contrasts a set of genomic variations with the
information contained in a database that follows the
Conceptual Model of the Human Genome (CMHG)
(Reyes Román et. al., 2016).
Applying GeIS to the bioinformatics domain is a
fundamental requirement, since it allows us to
structure the Human Genome Database (HGDB)
with “curated” and “validated” data. The initial
research on applying CM approaches to the human
genome was reported in the works of Paton
(Bornberg-Bauer and Paton, 2002) and Ram (Ram
and Wei, 2004). The main goal in Ram's work was
to show the advantages and benefits of using
conceptual modeling to compare and search for the
protein in 3D (see other related works in (Pastor et.
al., 2010)). Reyes et. al. (2016) describes a CMHG
which proposes a domain definition at the
conceptual level. From this CMHG, we generated a
GeIS to support VarSearch. The application of CM
helps us to better understand and manage the
knowledge of the human genome.
This paper is divided as follows: Section 2 describes
the design and implementation of the VarSearch tool
as a GeIS. Section 3 describes two case studies
carried out using VarSearch. Finally, Section 4
contains our conclusions and outlines future work.
2 DESIGN AND
IMPLEMENTATION OF A GeIS
A GeIS can be defined as a system that collects,
stores, manages and distributes information related
to the behavior of the human genome. As mentioned
above, the GeIS described here is based on the
CMHG. This section deals with the VarSearch
design stage.
2.1 Design Overview
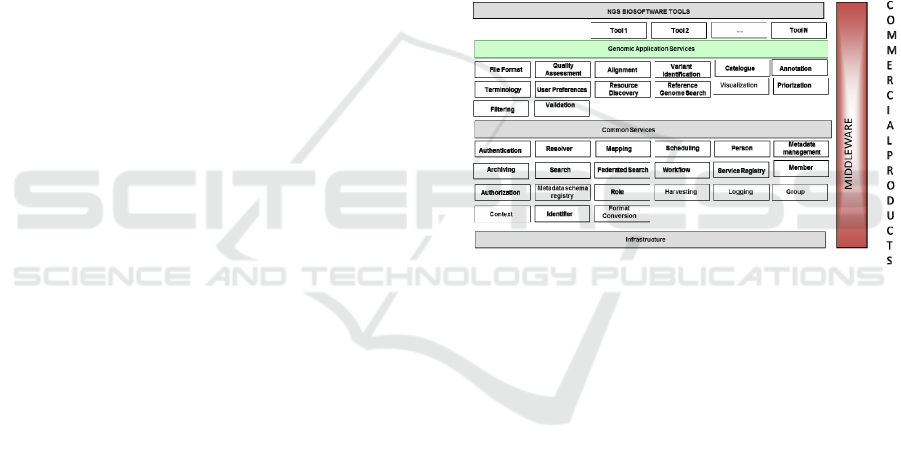
As outlined at Figure 1, VarSearch is based upon the
E-Genomic Framework (EGF), described in depth in
different research papers such as (Roldán et. al.,
2014). For the implementation of the tool, a series
of steps were carried out to ensure its good
performance, as explained below:
a) Human Genome Database (HGDB): The
transformation of the model defined for the database
schema (logical model) was almost automatic, using
the Moskitt tool (Muñoz et. al., 2010). In this task,
we found two different levels of abstraction in the
model. The conceptual model represents the domain
from the point of view of scientific knowledge,
while the database schema focuses on the efficient
storage and retrieval of data. For this reason, the
details of the physical representation must be
considered to improve the final implementation. It is
important to emphasize the integration of the two
tables "Validation" and "Curator" in the DB schema.
These tables are not actually part of the knowledge
representation of the domain, but are necessary for
the development and implementation of the tool.
Figure 1: EGF Framework and VarSearch (Roldán et. al.,
2014).
To load the HGDB the SILE methodology
(Reyes and Pastor, 2016) was used, which was
developed to improve the loading processes and
guarantee the treatment of "curated data". SILE was
used to perform the “search” and “identification” of
variations associated with a specific disease (a task
validated by experts in the genetic domain). When
the identified and curated data have been obtained
the "selective loading” is performed (through the
loading module) in the HGDB. The data loaded are
then “exploited” by VarSearch. Some of the diseases
(of genetic origin) studied and loaded were Alcohol
Sensitivity, Neuroblastoma, etcetera.
b) Selection of the different data sources: For
the choice of data sources, we addressed the
requirements raised in this first phase of the project.
After conducting studies and analysis of various
genomic repositories, we selected the following
databases: NCBI, dbSNP, UMD and BIC.
NCBI (https://www.ncbi.nlm.nih.gov/) is a data
source with curated data on structural concepts of
VarSearch: Annotating Variations using an e-Genomics Framework
329
DNA sequencing. From this repository, we extracted
information related to chromosomes, genes,
transcripts, exons... and everything related to the
"Structural view" of the CMHG. dbSNP (Sherry et.
al., 2001), BIC (Szabo et. al., 2000) and UMD
(Béroud et. al., 2000) are databases of variations that
store curated information on genetic differences
between individuals. The main reason for using
dbSNP is because it not only focuses on variations
of a specific gene or region, but also contains
variations related to all chromosomes and updates
the information immediately. BIC and UMD were
selected because of the requirements of a research
group that was collaborating with us in a project
(Future Clinic Project) focused on "breast cancer".
This group helped us to test the performance of the
GeIS and its associated tool.
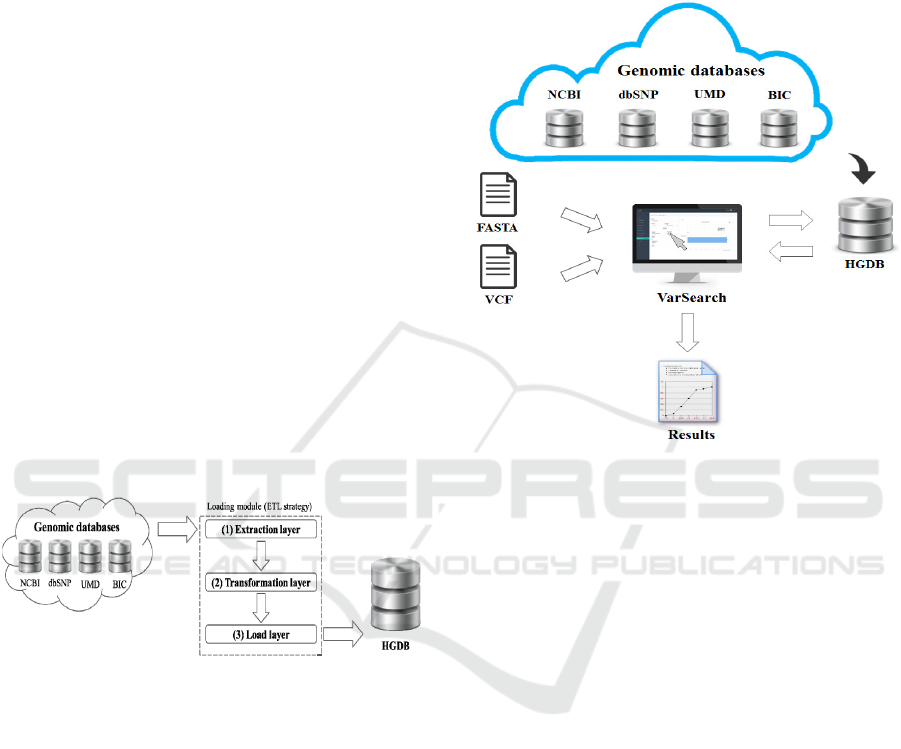
c) Genetic loading module: For the loading
process of the HGDB, a load module was designed
to store the data from the previously measured data
sources. This load module was developed using an
ETL strategy (Zhou et. al., 2011) with three different
levels: extraction, transformation, and load (see
Figure 2). Each level is completely independent of
the others, facilitating and clarifying the design of
the system and improving its flexibility and
scalability.
Figure 2: Load module.
As can be seen in Figure 2, all the necessary
information is extracted from the source databases in
the first layer (1). All this raw unstructured data goes
to the second layer (2) where several transformations
are made to format the data according to the structure
of the database schema. These transformed data are
sent to the third layer (3), which communicates
directly with the database (following the above-
mentioned SILE methodology in Task “a”).
2.2 VarSearch Tool
VarSearch is a web application that allows the
analysis of variations obtained from the DNA
sequenciation of biological samples and which is
stored in FASTA or VCF file formats (Claverie and
Notredame, 2011). Different users can access the
application in private spaces in the HGDB and each
user can address his own variations. The validation
of variations that they consider relevant can be
included. It also offers storage for the users’
variations in order to find similarities in the file
analysis process.
Figure 3: VarSearch application.
Another advantage is the inclusion of the
information obtained from the data sources, together
with the user validations in the database, which is an
improvement in performance related to the search
for variations. VarSearch is capable of finding
variations in the database from a provided file (see
Figure 3). The variations found are displayed to the
user, and any additional information that the file
lacks can be calculated and validated. Any variations
of the file that have not been found in the database
can also be stored. After inserting one or more
variations not found in a file -because they are
considered relevant to the user- and reanalysing this
file, these inserted variations will be found in the
database and displayed to the user.
The VarSearch functionality has been grouped
into three main packages: (a) User management: a
user can act as administrator and control other users,
or can create new users and modify or eliminate
their Information. (b) Data load management: the
system allows the user to load the files to be
analysed in both VCF and FASTA format (Agliata
et. al., 2014), compare the variations in these files to
the variations in the HGDB used by VarSearch. (c)
Data analysis: After analysing and verifying the
variations in the input files, the user can list the
variations and classify them by multiple criteria
ENASE 2018 - 13th International Conference on Evaluation of Novel Approaches to Software Engineering
330

(position, chromosome, existence in the database,
etc.). There is also a series of functionalities related
to the login and modification of account information
that has not been grouped in any functionality
package.
2.2.1 Confidentiality of the Information
As this information is a company’s primary
resource, VarSearch restricts access to it. When a
user validates a variation, he can choose a privacy
category: (a) Public content, if he is willing to share
the knowledge with other users, or (b) Private
content, allowing access only to the owner-user and
hidden from other users. All the variations can only
be accessed by the user who created them.
2.2.2 VarSearch Architecture
In order to make it accessible to all users, VarSearch
was designed as a web application with HTML5
technology in a language common to all current
browsers. The information is managed by the
MySQL database. The VarSearch architecture
consists of the following elements:
(a) A distributable database based on MySQL
(using software tools like: Navicat Enterprise
and MySQL Workbench). For the initial
validation of this database, we only loaded the
information related to chromosomes 13 and 22.
(b) A set of REST services (Haupt et. al., 2014)
developed in Java using Hibernate and Jersey,
which are deployed on a Tomcat server 7.
(c) A web application, which uses the Bootstrap
framework for general organization of the
interface and files, together with jQuery to
define advanced interface components and
invoke REST services.
(d) It also includes a "mini" REST service to
manage users and roles, which is based on the
same architecture and technologies as the
other REST services. The data layer is based
solely on MySQL (you can see the VarSearch
architecture represented in Figure 4).
The application entry point is a file with
variations detected by a sequencing machine in VCF
or FASTA format. With this input the database is
searched to detect any variations, additional
information on the diseases they may cause and the
associated bibliography.
VarSearch users follow this process when
working with the tool:
• A VCF file is uploaded from the web.
Figure 4: VarSearch Architecture.
• The file is then processed and parsed. The
entries are shown on an HTML table and the
variants of each VCF entry can be seen.
• The variations present in the input file can be
annotated against the database and the
annotated file is downloaded in *.xls, *.csv
and *.pdf format or its contents viewed in
another HTML table.
To parse the VCF file and annotate the variants,
VarSearch relies on snpEff and snpSift (Tolhuis and
Wesselink, 2015) tools, and so well tested libraries
are used instead of reinventing the wheel. This also
ensures VCF standard support, using ANN files for
variant annotation. If another type of information is
considered useful for annotation and not covered by
the procedure described, VarSearch uses the “INFO”
field to introduce the desired values. As VarSearch
is based on EGF, new genome annotation files can
be quickly integrated by developing the proper
parser module, either by a custom development or
integrating a third-party tool or library.
All the information associated with the variations
found in the HGDB can be obtained. For variations
in the lists, user validations can be integrated for
future searches with the "Add Validation" option.
Another advantage of VarSearch is user
management (new users can be created and edited
using the "User Management" option). One of the
objectives of VarSearch is to continue the extension
and implementation of all the knowledge defined in
the CMHG, such as the treatment of pathways and
metabolic routes (Reyes Román et. al., 2017). This
tool facilitates the analysis and search for variations,
improving the generation of genomic diagnoses
associated with diseases of genetic origin. End-users
will find the web application easy to use and they
are guaranteed security for their data.
VarSearch: Annotating Variations using an e-Genomics Framework
331
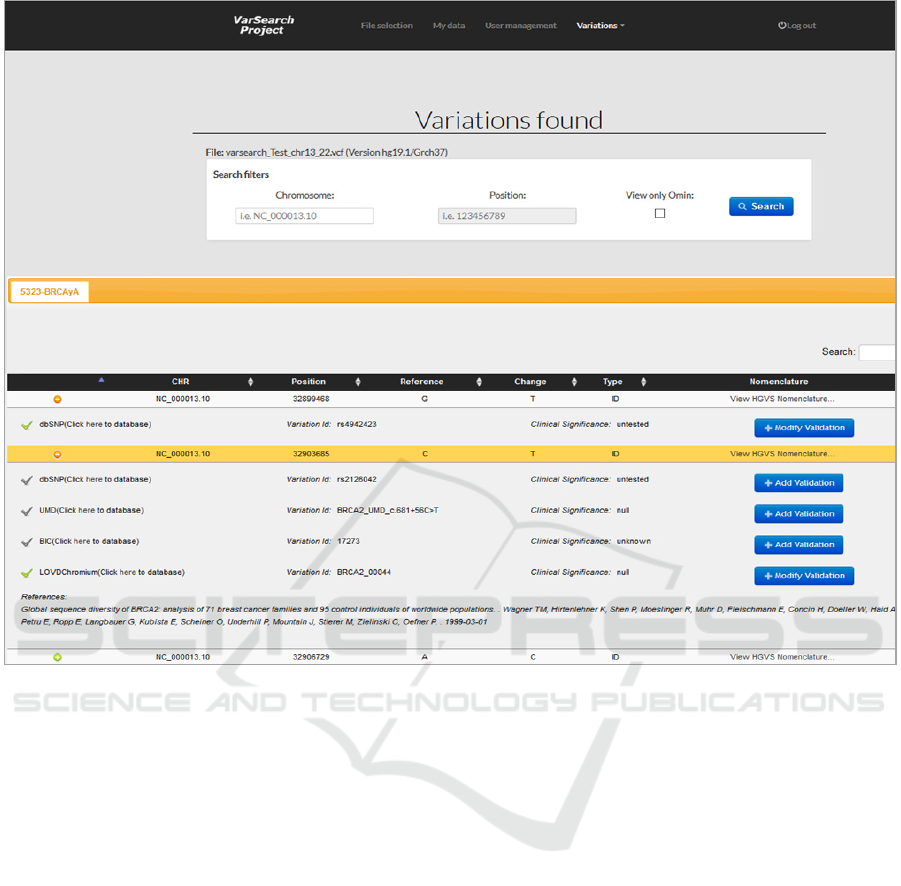
Figure 5: Analysis of VCF file using VarSearch.
3 CASE STUDIES USING
VARSEARCH
To verify VarSearch performance, two case studies
were carried out. In the first, VarSearch was used to
analyse a VCF file. The second compared the time
spent on searching for variations manually and using
the application. To access the application VarSearch
users must have an account provided by Gembiosoft
SME (http://gembiosoft.com/). After logging in, a
file is selected for analysis. VarSearch reads all
records and transforms them into variations. These
transformations depend on the file information: for
example, the FASTA files contain a genetic
sequence (NG), and so require the reference on
which the variation is based to be to the “NG”
sequence. In contrast, VCF files use positions
relative to chromosomes (“NC”). Once the file
records have been converted into variations, the next
step is to search for these variations in the HGDB.
After the analysis, the “variations found” and
“variations not found” can be differentiated.
• Found Variations Management: Found
variations are those extracted from the file in
which information has been found in the
HGDB, which means that this variation has
been found in at least one genomic repository.
A found variation has much more information
than the variation obtained from the file and
allows us to calculate and submit detailed
information to the user. Having analysed the
VCF file, all the variations found are
displayed to the user, in each case calculating
the HGVS notation, its data source identifier,
clinical significance, and the number of
validations and databases found together with
their bibliographic references.
This information is calculated for VCF and
FASTA; however, VCF variations are sorted
by samples. Figure 5 shows the results
obtained by analysing a VCF file with a single
sample. For this sample (5323-BRCAyA), a
ENASE 2018 - 13th International Conference on Evaluation of Novel Approaches to Software Engineering
332
number of variations were found with the
corresponding information. A variation can
have validations made by users.
The validation column corresponds to the
number of validations that each variation has
and if a validation is private, only the owner
will see it. Another VarSearch feature is its
support for multiple bibliographical
references. A variation can be found in
different DBs and may contain different
bibliographic references.
• Not found Variations (insertion and treatment):
The user who is analysing variations may find
a variation in the file, which was not found in
the database. Using his experience and
knowledge he may consider some variations
as relevant despite not being found.
With VarSearch the user can insert the not
found variations or any variation considered
key to the study. If the user has inserted
certain variations that had not been found, on
reanalysing the file these inserted variations
are compared with the variations in the file,
showing the similarities. To differentiate the
variations of the different repositories from
user variations, the results obtained from the
user’s experience and the results from years of
study of different biomedical databases are
differentiated.
3.1 Improved Efficiency and Time in
Finding Variations with VarSearch
To validate the effectiveness and performance of the
proposed software, some experiments were
performed to measure efficiency and time. A study
was conducted to compare the time spent searching
for variations manually with an automatic search of
all the repositories mentioned above using
VarSearch.
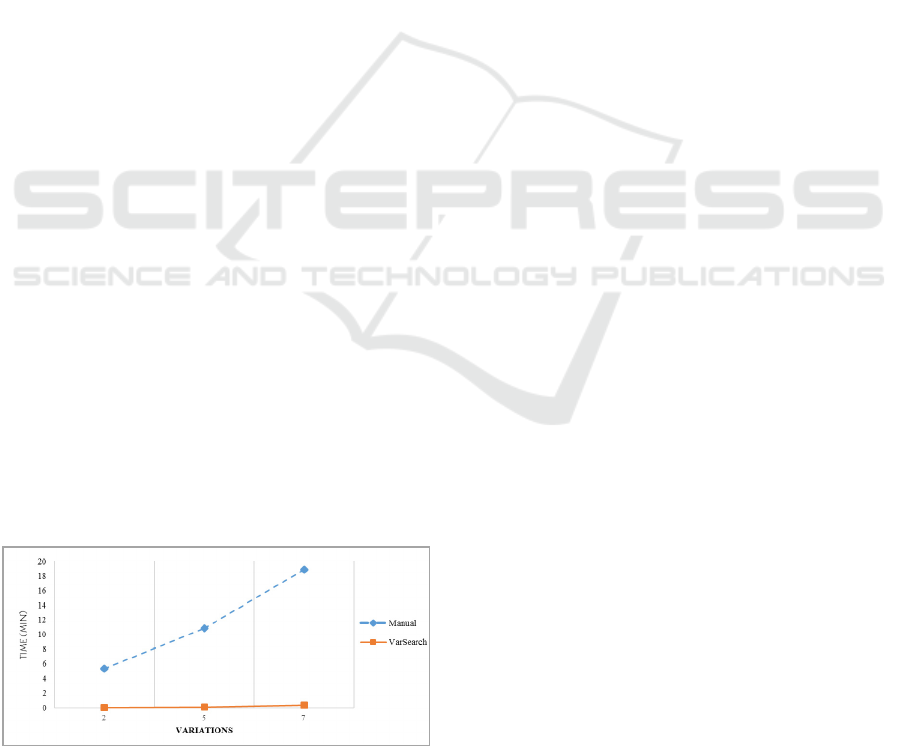
Figure 6: Time optimization.
A manual search of one variation involves
detecting the variation in the VCF or FASTA file, a
search for the variation in the different databases,
and the identification and verification of the
variation. VarSearch was tested for the time it
needed to search for several variations, calculating
the time evolution according to the number of
variations involved (2, 5 and 7). The results can be
seen in Figure 6.
As can be seen in Figure 7, the cost of
performing a manual search rises to 5’32 minutes for
2 variations, 10’83 minutes for 5 variations and
18’89 minutes for 7 variations. However, with
VarSearch the time remains constant at between 2
and 3 seconds for different variations, which
confirms its efficient performance. Using this tool
thus significantly reduces the time spent on the
search for variations. Also, it must be remembered
that the manual search process does not calculate
additional information for variations. If this
information were necessary, the search time would
increase significantly, however, with VarSearch this
time remains constant because this information has
already been calculated in the search for variations.
4 CONCLUSIONS
VarSearch is a flexible new analysis framework or
web application that provides a powerful resource
for exploring both “coding” and “non-coding”
genetic variations. To do this, VarSearch integrates
VCF format input/output with an expanding set of
genome information. VarSearch (and other tools
built on EGF) will therefore facilitate research into
the genetic basis of human diseases.
EGF can also be expected to allow the
development of new tools in diverse e-genomics
contexts. As genetic laboratories are now oriented to
facilitating genetic procedures, web access, usability
and feasibility, the definition of different profiles are
therefore important goals. All this allows the user to
configure the tool according to his specific needs.
These necessities include inserting genetic variations
and validating its own variations, thus increasing its
“know-how”. VarSearch was tested in two different
case studies; one focused on the analysis of
variations (insert and search) and another to test its
search performance.
Future work will be oriented to the
implementation of -haplotypes and statistical
factors- (i.e., frequencies and populations) and
improving the next version of VarSearch (prototype)
for genetic diagnosis. Future research work will also
VarSearch: Annotating Variations using an e-Genomics Framework
333

be aimed at the application of Data Quality (DQ)
metrics to enhance the HGDB. We also intend to
extend the model with studies on the treatment of
“haplogroups”, including subjects with a similar
genetic profile who share a common ancestor.
ACKNOWLEDGEMENTS
This work was supported by the Generalitat
Valenciana through project IDEO
(PROMETEOII/2014/039) and the Spanish Ministry
of Science and Innovation through Project DataME
(ref: TIN2016-80811-P). The authors are grateful to
Jorge Guerola, Francisco Valverde, Ana León
Palacio, Ainoha Martín, Verónica Burriel Coll,
Mercedes Fernández A., Carlos Iñiguez-Jarrín,
Lenin Javier Serrano and Ma. José Villanueva for
their valuable assistance.
REFERENCES
Mardis, E. R., 2008. Next-generation DNA sequencing
methods. In Annu. Rev. Genomics Hum. Genet., vol. 9,
pp. 387-402, doi: 10.1146/annurev.genom.9.081307.
164359.
Grosso, L. A., 2016. Precision medicine and cardio-
vascular diseases. In Rev Colomb Cardiol, vol. 23, no.
2, pp. 73-76, doi: http://dx.doi.org/10.1016/j.rccar.
2016.01.026.
Olivé, A., 2007. Conceptual modeling of information
systems. Springer-Verlag Berlin Heidelberg, pp. 1-
445, doi: 10.1007/978-3-540-39390-0.
Reyes Román, J. F. et. al., 2018. Genomic Tools*: Web-
applications based on Conceptual Models for the
Genomic Diagnosis. In selected papers from ENASE
2017 in Communications in Computer and
Information Science (CCIS). Springer, pp. 1-21.
Reyes Román, J. F. et. al., 2016. Applying Conceptual
Modeling to Better Understand the Human Genome.
In Comyn-Wattiau I., Tanaka K., Song IY., Yamamoto
S., Saeki M. (eds) Conceptual Modeling. ER 2016.
Springer International Publishing, pp. 404-412, doi:
10.1007/978-3-319-46397-1_31
Bornberg-Bauer, E. and Paton, N. W., 2002. Conceptual
data modelling for bioinformatics. In Briefings in
Bioinformatics, vol. 3, no. 2, pp. 166-180, doi:
10.1093/bib/3.2.166.
Ram, S. and Wei, W., 2004. Modeling the semantics of 3D
protein structures. In Conceptual Modeling–ER 2004,
Proceedings. pp. 696-708, doi: 10.1007/978-3-540-
30464-7_52.
Pastor, M. A. et. al., 2010. Conceptual Modeling of
Human Genome Mutations: A Dichotomy Between
What we Have and What we Should Have. In
BIOSTEC Bioinformatics 2010, pp. 160-166, ISBN:
978-989-674-019-1.
Roldán M., D. et. al., 2014. An integration architecture
framework for e-genomics services. In 2014 IEEE
Eighth International Conference on Research
Challenges in Information Science (RCIS), doi:
10.1109/RCIS.2014.6861063.
Muñoz, J. et. al., 2010. Configuring ATL transformations
in MOSKitt. In Proceedings of the 2nd. International
Workshop on Model Transformation with ATL (MtATL
2010). CEUR Workshop Proceedings.
Reyes Román, J.F. and Pastor, O., 2016. Use of GeIS for
Early Diagnosis of Alcohol Sensitivity. In
Proceedings of the 9th International Joint Conference
on Biomedical Engineering Systems and Technologies,
vol. 3, pp. 284-289, doi: 10.5220/0005822902840289.
Sherry, S. T. et. al., 2001. dbSNP: the NCBI database of
genetic variation. In Nucleic acids research, vol. 29,
no. 1, pp. 308-311.
Szabo, C. et. al., 2000. The breast cancer information core:
database design, structure, and scope. In Human
mutation, vol. 16, no. 2, pp. 123, doi: 10.1002/1098-
1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y.
Béroud, C. et. al., 2000. UMD (Universal mutation
database): a generic software to build and analyze
locus-specific databases. In Human mutation, vol. 15,
no. 1, pp. 86, doi: 10.1002/(SICI)1098-1004(200001)
15:1<86::AID-HUMU16>3.0.CO;2-4.
Zhou, H. et. al., 2011. An ETL strategy for real-time data
warehouse. In Practical applications of intelligent
systems, Springer Berlin Heidelberg. pp. 329-336, doi:
https://doi.org/10.1007/978-3-642-25658-5_41.
Claverie, J. M. and Notredame, C., 2011. Bioinformatics
for dummies. In John Wiley & Sons, pp. 1-456, ISBN:
978-0-470-08985-9.
Agliata A. et. al., 2014. IGV-plus: A Java Software for the
Analysis and Visualization of Next-Generation
Sequencing Data. In Vogiatzis C., Walteros J.,
Pardalos P. (eds) Dynamics of Information Systems.
Springer Proceedings in Mathematics & Statistics, vol
105. Springer, Cham, pp 149-160, doi: https://doi.org/
10.1007/978-3-319-10046-3_8
Haupt, F. et. al., 2014. A model-driven approach for REST
compliant services. In IEEE International Conference
on Web Services (ICWS), pp. 129-136, doi: 10.1109/
ICWS.2014.30.
Tolhuis, B. and Wesselink, J. J., 2015. NA12878 Platinum
Genome GENALICE MAP analysis report.
Reyes Román, J. F. et. al., 2017. Software Engineering
and Genomics: The Two Sides of the Same Coin?. In
Proceedings of the 12th International Conference on
Evaluation of Novel Approaches to Software
Engineering (ENASE 2017), pp. 301-307, doi:
10.5220/0006368203010307.
ENASE 2018 - 13th International Conference on Evaluation of Novel Approaches to Software Engineering
334
