
Identifying a Medical Department based on Unstructured Data
A Big Data Application in Healthcare
Veena Bansal
1
, Abhishek Poddar
2
and R. Ghosh-Roy
3
1
Indian Institute of Technology Bhilai, Raipur, India
2
Indian Institute of Technology Kanpur, India
3
IBM UK Limited, London, U.K.
Keywords: Healthcare, Big Data, Unstructured Data, Tertiary Healthcare.
Abstract: Health is an individual’s most precious asset and healthcare is one of the vehicles for preserving it. The Indian
government’s spend on healthcare system is relatively low (1.2% of GDP). Consequently, Secondary and
Tertiary government healthcare centers in India (that are presumed to be of above average ratings) are always
crowded. In Tertiary healthcare centers, like AIIMS, patients are often unable to articulate correctly their
problems to the healthcare center’s Reception staff for these patients to be directed to the correct healthcare
department. In this paper, we propose a system based on Big Data and Machine Learning to direct the patient
to the most relevant department .We have implemented and tested parts of this system wherein a patient enters
his symptoms and/or provisional diagnosis; the system suggests a department based on this user input. Our
system suggests the correct department 68.05% of the time. Our system presently makes its suggestions using
gradient boosting algorithm that has been trained using two information repositories- symptoms and disease
data, functional description of each medical department. It is our informed assumption that, once we have
incorporated medicine information and diagnostics imaging data to train the system and the complete medical
history of the patient, performance of the system will improve significantly.
1 INTRODUCTION
Everyone strives to be healthy and stay away from
hospitals but occasionally one must visit a healthcare
facility. Healthcare in India is a three-tier system;
Primary care is the first line of contact, often between
a patient and a doctor. Secondary and Tertiary
healthcare centers require a referral from a Primary
healthcare center. Tertiary healthcare centers cater for
complicated medical conditions and require
specialized medical consultations.
A sample referral is shown in Figure 1. The
referral has the name of a patient, provisional
diagnosis and the hospital name to which the patient
has been referred to but more often without the details
of the department within the hospital. The Tertiary
healthcare centers such as AIIMS (All India Institute
of Medical Science) have multiple departments, with
near unique capabilities in each department for
treating ailments. Even medically literate patients
often have difficulty in identifying the correct
department. The healthcare center’s Reception staff is
often the first port of call and these staff often quickly
browse through the medical documents of a patient to
identify the appropriate department; this is not fool
proof and mistakes are often made, leading to
inconveniences caused downstream to all parties
concerned. This is a major bottleneck, especially as
the system must deal with many thousands of patients
each day.
People who have access to the Internet, and have
the required skill sets, can collate information about
each department before making an online
appointment. However, for many people in India,
they do not even have access to the Internet and/or not
literate enough to make an online appointment.
Irrespective of the channel used for booking, all
walk-in patients face very similar challenges of
identifying the correct department to proceed to. We
have therefore focused on the walk-in process where
most of the errors have been noticed. It was our
conclusion that we need to first augment the manual
appointment booking process to identify the correct
department, thereby make the overall booking
process easier and error free for the patients. In this
work, we propose a system that will automatically
Bansal, V., Poddar, A. and Ghosh-Roy, R.
Identifying a Medical Department based on Unstructured Data.
DOI: 10.5220/0006773904750482
In Proceedings of the 20th International Conference on Enterprise Information Systems (ICEIS 2018), pages 475-482
ISBN: 978-989-758-298-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
475

Figure 1: A Sample referral to a tertiary healthcare system.
recommend an appropriate department to the patients
by looking at their medical documents.
We have reviewed the related work in §2 and
presented a formal model of our proposed system in
§3. An implementation of our system has been
detailed in §4. Results and conclusions have been
presented in §5.
2 RELATED WORK
Over the years, several computational systems for
decision making have been used in healthcare. These
have either helped humans in reducing their workload
or helped in decision making or both. Expert systems
built to diagnose a disease (Naser et al, 2010; Tenorio,
2011; Rahman and Hossain, 2013; Ibrahim, 2014)
have faced a challenge in clearly representing medical
history of a patient. Supervised learning techniques
such as decision trees, Bayesian classifiers, artificial
neural networks, support vector machines and k-
nearest neighbors have also been used in building
expert systems. A decision support system can also be
rules or fuzzy rules based (Rahman and Hossian,
2013). These systems are used for diagnosing the
presence of a disease, or predicting adverse effect of
a drug (Fosamax), or predicting the onset of a disease
(Ibrahim, 2014; Ephzibah and Sundarapandian, 2012;
Jain and Raheja, 2015).
Another line of research led to the development of
systems that helped patients in managing their diet
and medicines (Caballero-Ruiz et al, 2017; Goethe
and Bronzino, 1995); some helped Health Insurance
Providers with pre-authorization of insurance
requests (Araújo, 2016); others helped doctors in
identifying the best possible treatment for a given
disease (Delias, 2015) or even recommending
pathological tests (Alonso-Amo, 1995); or check the
efficacy of an ongoing treatment (McAndrew, 1996).
All these systems require a vast amount of data
(Davenport, 2014) and with the advent of Big Data
(Aruna Sri and Anusha, 2016), a new set of
possibilities in healthcare have emerged (Schultz,
2013). Prevention strategies and treatment
recommendations are all based on vast amount of data
(Saravan, 2015). The medical world has not yet
evolved a standard terminology to describe medical
conditions and medical departments. Work is being
carried out to create a standard medical language to
be used across applications and platforms
(Handerson, 2016).
We extensively searched for an application or a
system that can provide a description of all diseases
and respective departments of hospitals that treat
these diseases. To the best of our knowledge, no such
application exists. Such an application or a system
can help a patient identify the appropriate department
of a hospital for a specific treatment. We spoke with
the doctors in Secondary and Tertiary healthcare
facilities, and they all confirmed that patients are
often directed to the wrong department by the
Reception staff. Sometimes, patients are not even able
to describe their problems. Often the Reception staff
are unable to decipher the medical reports/documents
provided by the patients. A patient often therefore
ends up wasting his own time; the hospital also ends
up wasting its own resources if the patient ends up at
the wrong department. We have, hence, decided to
build a system that will direct patients to the most
appropriate department of the healthcare facility. Our
system is based on Big Data techniques and is
described next.
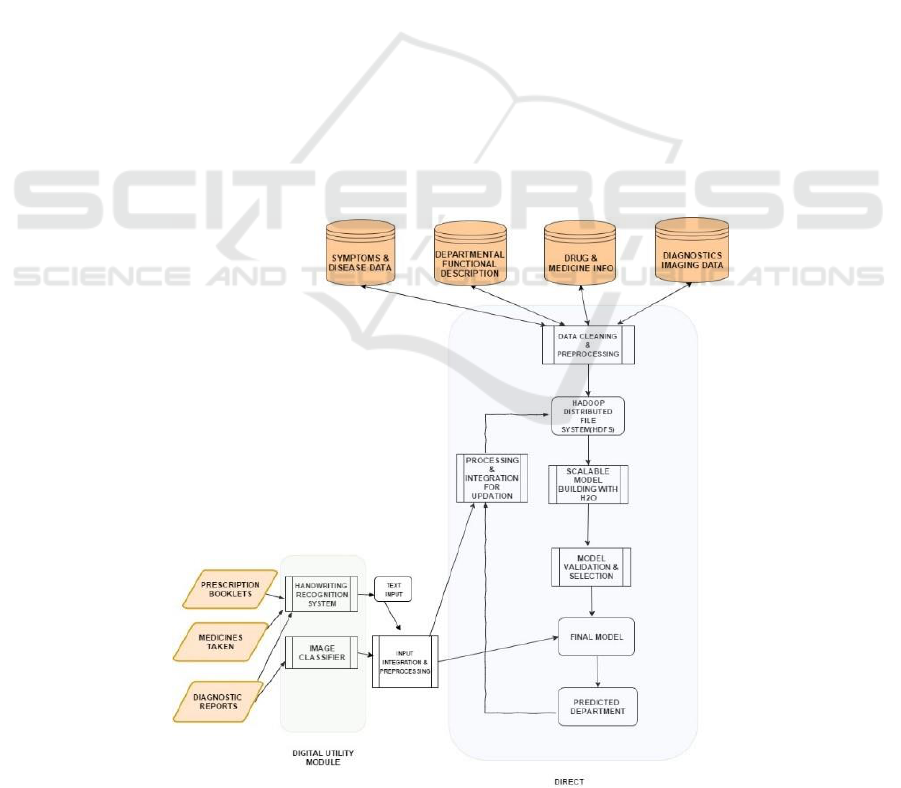
3 THE PROPOSED MODEL
The block diagram of our proposed system is given in
Figure 2. When patients walk into a Tertiary
healthcare center, their documents can be scanned
including:
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
476

previous prescriptions from doctors
medicines taken
diagnostic reports & medical images
The scanner would then digitize the documents.
The digitized documents would then be used by the
Digital Utility Module (referred to as DUM). The
DUM would pre-process and extract the information
presented in the documents and images. The images
will then be processed, extract metadata if available,
to identify the organs and other relevant details
available in the images (Filipovych and Davatzikos,
2001; Kucheryavski, 2007; Antoio at al, 2001).
Prescriptions, reports, bills etc. will be processed by
OCR and ICR engines to convert them into searchable
and editable text (Ciregan, 2012; Patel et al, 2012).
The extracted information will then be passed to
the next module, called DIRECT. The DIRECT
module will recommend a hospital department based
on the input. DIRECT is at the heart of our system,
has the knowledge base and processes the inputs
provided by patients to recommend an appropriate
department. DIRECT employs a machine learning
model that is trained and validated offline. The
training process involves the following steps that we
explain next.
Data Cleaning & Preprocessing
Scalable Model Building
Model Validation & Selection
Preprocessing & integration for updates
Data Cleaning and Preprocessing
We need a labeled dataset to train the system. For
instance, we can train the system to learn the disease
that each hospital department treats by using data
containing diseases mapped to an appropriate hospital
department. This process includes creating a profile
for each disease based on its symptoms, medicines,
and diagnostic reports and then mapping each disease
profile to a department. This includes extracting the
useful parts of the text, purging the stop-words from
the text (Ullman and Rajaraman, 2011), converting
the words into a common form by using stemming
(Lovins, 1968), feature extraction from the texts
(Guyon and Elisseff, 2003) and converting the data
into a vector space model (Ripley, 1996). The
challenging part of the problem is that apart from text
data, there are also image data to deal with. According
to data types, we have loosely three classes of
extracted features – the symptoms or disease name,
the medicines taken and processed images. While
training, the model will learn to assign weights to
each class of features.
Scalable Model Building
Gradient Boosting Machine (Click et al, 2017), Deep
Learning (a multi-layer neural network model
trained using back-propagation algorithm) (Candel
et al, 2015) and Distributed Random Forest (H2O
website) are the models that we have selected based
on their potential and performance. Our main task is
multinomial classification (Aly, 2005).
Model Validation & Selection
This is the process where we select a final model
based on varying criteria like log loss (Collier, 2015)
or misclassification rates among all the different
models. There are many hyper parameters in each
machine learning model that get tuned during this
training.
Processing & Integration Updation
This process of the DIRECT module will enhance the
accuracy of the system over time as it sees and learns
from more and more real use cases. The predicted
department and the inputs from the patients are pre-
processed in the agreed format of our training data. It
is then added to our knowledge base for continuous
learning.
4 SYSTEM IMPLEMENTATION
We have implemented part of the proposed system
called The Tertiary Healthcare Center Directing
System. The complete system needs four information
repositories for training: Symptoms & Disease Data
(names of diseases and their symptoms),
Departmental Functional Description, Drug &
Medicine Information, Diagnostics Imaging Data.
The form of Symptoms & Disease data is as follows.
<symptom
1
, symptom
2
, …, symptom
n
> <disease
1
>
Functional description of each department is
represented as follows.
<disease
1
, disease
2
, …, disease
m
><medical_deptt>
Drug & Medicine information consists of the
following information.
<drug
1
, drug
2
, …, drug
k
><disease
1
>
Diagnostics and Imaging data has two
components: Image and corresponding diagnosis. We
have used Symptoms and Disease data as well as
Departmental Functional Descriptions to train and
test the system. We have not yet incorporated Drug
and Medicines Information, Diagnostics Imaging
data.
Identifying a Medical Department based on Unstructured Data
477
The data for training and validating the system is
not available in the required form and requires pre-
processing. Hence, the system implementation
includes the following phases:
a. Finding a dataset that has disease
information (possibly including their
names, associated symptoms, types,
synonyms etc.) and name of the concerned
medical department.
b. Converting the above dataset into vectors.
c. Identify suitable machine learning models
and train them.
d. Test the models and select the best
performing model.
We created a labeled dataset using a disease-
description dataset and a document on functional
descriptions of hospital departments using heuristics.
We used dataset from the Disease Ontology project
called doid-non-classified.obo (DOID, 2017)
(referred to as disease_description). Each disease has
an assigned identifier, name, symptoms and some
other details. We also created functional descriptions
(referred to as functional_description) of healthcare
departments from two different sources (Henderson,
2016; Mayoclinic Website, 2017). The datasets
disease_description and functional_description
contain information about 10612 diseases and 20
healthcare departments respectively. The system
compares each disease description with all the
departmental descriptions to assign each disease to a
department. This is a challenging task and involves
text processing. We had to remove stop words,
perform stemming, used heuristics to handle
synonymous, homonymous, etc. For instance, the pair
of words electrocardiogram and cardiomyopathy are
essentially the same whereas hypertension and
hyperbola are totally unrelated. We used python to
implement the preprocessing phase of the system.
We obtained a labeled dataset where each disease
is mapped to a hospital department. The dataset is
converted into a vector space model using term-
frequency-index and document-frequency technique
(tfidf). The labelled dataset presented as vectors have
been used to train and test machine learning models.
Our problem is essentially a multinomial
classification task (Aly, 2005). We had department
names as our classes and the objective of our model
was to learn a mapping between disease descriptions
and departments. We split the datasets into two parts:
65% for training, 35% for testing. We trained three
machine learning algorithms: Gradient Boosting
Machine, Distributed Random Forest and Deep
Learning. We implemented the system using an open-
source big data analysis platform (H2O, 2016).
Figure 2: The block diagram of our system.
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
478

Table 1: List of medical departments in a hospital.
Serial
No.
Department Name
1
Anesthetics
2
Breast Screening
3
Cardiology
4
Ear, nose and throat (ENT)
5
Elderly services department
6
Gastroenterology
7
General Surgery
8
Gynecology
9
Hematology
10
Neonatal Unit
11
Neurology
12
Nutrition and dietetics
13
Obstetrics and gynecology
units
14
Oncology
15
Ophthalmology
16
Orthopedics
17
Physiotherapy
18
Renal Unit
19
Sexual Health
20
Urology
We choose the model with the lowest
misclassification rate as our final model. The results
from our model building, validation and selection
phase have been discussed in the next section.
5 RESULTS AND DISCUSSIONS
We have used three machine learning models, namely
Gradient Boosting Machine, Deep Learning and
Distributed Random Forest. Each of these models
require learning that essentially amounts to tuning
some of the hyper parameters.
Random Forest hyper parameters include the total
number of trees to grow, maximum tree depth and the
number of predictors randomly sampled as candidates
for each split.
Neural networks have variants such as hyperbolic
tangent (Kalman and Kwasny, 1992), rectifier
(Hahnloser et al 2000) and maxout (Goodfellow,
2013); each of these could optionally be paired with a
regularization technique called dropout (Srivastava et
al, 2014). There are many hyper-parameters to be
tuned (Han and Kamber, 2001).
Hyper parameters of Gradient Boosting that need
tuning include the number of trees to be constructed,
the maximum depth of each tree, percentage of rows
to be sampled per tree, and learning rate. There are
certain guidelines for tuning these parameters
(Friedman, 1999; 2002).
Table 2 summarizes the results that we have
obtained from these three models. We trained all three
machine learning models using 5 different settings of
the parameters. After training the system, we tested
using the same data. Column 2, 4 and 6 of Table 2
show false positive or misclassification for all 5
parameters settings for all three machine learning
models on the training data. We then tested the three
models with 5 different parameters settings using the
validation data which is new to the models. The
percentage of false positive is shown in columns 3, 5
and 7. The misclassification or false positives have
been plotted for better perception of the three models
with 5 different settings of hyper parameters and
shown in Figure 3. As mentioned in the previous
section, we have used 10,612 disease mapped to 20
hospital departments. It is obvious from the results
that, using just the descriptions of departments and
diseases, the system is able to suggest correct
department 89.82% of the time using Distributed
Random Forest on the training data. However, when
we run Distributed Random Forest on validation data,
it is able to suggest the correct department 60.19% of
the time only. Amongst all models and parameters
settings, the best validation performance is 68.05% of
Gradient Boosting Model. The performance across all
parameters settings and models is close to 70%.
It can therefore be concluded that the information
contained in our dataset cannot give us a performance
better than 70% true positives. Our system as shown
in Figure 2 has many other sources of information that
we need to incorporate for better performance as we
explain in the next section.
6 CONCLUSION AND FUTURE
WORK
We wanted to build a system that will help patients
going to tertiary health care system identify the
correct hospital department. We have implemented
and tested parts of this system wherein a patient enters
his symptoms and/or provisional diagnosis; the
system suggests a department based on this user
input. Our system suggests the correct department
68.05% of the time. To improve the performance
further, we need to incorporate medicine information
and diagnostics imaging data into our system as
shown in Figure 2. The system should take user’s past
prescriptions and diagnostic reports into account
when suggesting a medical department.
Identifying a Medical Department based on Unstructured Data
479

Table 2: Misclassification done by three different machine learning models with five different settings for hyper parameters
for training and validation data (GBM: Gradient Boosting Machine, DL: Deep Learning and DRF: Distributed Random Forest,
T: Training Data, V: Validation Data).
Parameters
Setting
G B M ( T )
G B M ( V )
D L ( T )
D L ( V )
D R F ( T )
D R F ( V )
1
21.94
39.03
34.80
35.55
15.38
34.14
2
51.34
55.04
33.42
34.52
15.02
32.91
3
30.40
43.74
32.90
33.96
12.14
33.57
4
17.72
31.95
32.43
33.78
10.99
35.96
5
13.82
39.35
33.3
33.47
10.18
39.81
Figure 3: Misclassification done by three different Machine Learning models for five different settings of hyper-parameters
with training and validation data; GBM: Gradient Boosting Method, DL: Deep Learning, DRF: Distributed Random Forest;
T: Training and V: Validation.
Once we incorporate everything, the performance
of this system will improve. We are now working on
integrating diagnostic image data. We have
experimented with image datasets of eyes and lungs.
We have been able to classify the organ in the image
with near 100% accuracy. We have yet to figure out a
mechanism to integrate the diagnostic image data into
the decision making process. We also want scan the
past prescriptions and run them through OCR/ICR to
convert them into text gain more information about
the treatment that the patient has received. Again, this
information must be integrated into decision making
process. We may have to map branded medicines into
generic medicines to be able to use this information
in the deciding the hospital department. Perhaps, our
knowledge base should contain a list of medicinal
compounds and the common diseases which they
treat, and a list of medicine names from different
brands for the same compound.
REFERENCES
S. Abu Naser, S. A., Al-Dahdooh R., Mushtaha, A. and El-
Naffar, M., 2010. Knowledge Management in ESMDA:
Expert System for Medical Diagnostic Assistance,
ICGST-AIML Journal, 10(1).
Josceli Maria Tenório, Anderson Diniz Hummel, Frederico
Molina Cohrs, and Vera Lucia Sdepanian, "Artificial
intelligence techniques applied to the development of a
decision–support system for diagnosing celiac disease,"
0
10
20
30
40
50
60
1 2 3 4 5
Error
Different Parameter Setting
GBM(T)
GBM(V)
DL(T)
DL(V)
DRF(T)
DRF(V)
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
480

International Journal of Medical Informatics, vol. 80,
no. 11, pp. 793-802, November 2011.
Saifur Rahaman and Mohammad Shahadat Hossain, "A
belief rule based clinical decision support system to
assess suspicion of heart failure from signs, symptoms
and risk factors," in 2013 International Conference on
Informatics, Electronics and Vision (ICIEV), Dhaka,
2013, pp. 1-6.
Neveen Ibrahim, Nahla Belal, and Osama Badawy, "Data
Mining Model to Predict Fosamax Adverse Events,"
International Journal of Computer and Information
Technology, vol. 3, no. 5, September 2014.
E.P. Ephzibah and Dr. V. Sundarapandian, "A NEURO
FUZZY EXPERT SYSTEM FOR HEART DISEASE
DIAGNOSIS," Computer Science & Engineering: An
International Journal (CSEIJ), vol. 2, no. 1, February
2012.
Vaishali Jain and Supriya Raheja, "Improving the
Prediction Rate of Diabetes using Fuzzy Expert
System," I.J. Information Technology and Computer
Science, vol. 7, no. 10, pp. 84-91, September 2015.
Estefanía Caballero-Ruiz et al., "A web-based clinical
decision support system for gestational diabetes:
Automatic diet prescription and detection of insulin
needs," International Journal of Medical Informatics,
vol. 102, pp. 35-49, June 2017.
Flávio H.D. Araújo, André M. Santana, and Pedro de A.
Santos Neto, "Using machine learning to support
healthcare professionals in making preauthorisation
decisions," International Journal of Medical
Informatics, vol. 94, pp. 1-7, October 2016.
Pavlos Delias, Michael Doumpos, Evangelos Grigoroudis,
Panagiotis Manolitzas, and Nikolaos Matsatsinis,
"Supporting healthcare management decisions via
robust clustering of event logs," Knowledge-Based
Systems, vol. 84, pp. 203-213, August 2015.
F. Alonso-Amo, A. Gomez Perez, G. Lopez Gomez, and C.
Montens, "An Expert System for Homeopathic
Glaucoma Treatment (SEHO)," Expert Systems with
Applications, vol. 8, no. 1, pp. 89-99, 1995.
Goethe, J. W. and Bronzino, J. D., 1995. An expert system
for monitoring psychiatric treatment, IEEE Engineering
in Medicine and Biology, 776-780.
McAndrew, P. D., Potash, D. L., Higgins, B., Wayand, J.
and Held, J., 1996. Expert system for providing
interactive assistance in solving problems such as
health care management , USPTO No. 5517405.
Thomas H. Davenport, Big Data at Work. Boston,
Massachusetts: Harvard Business Review Press, 2014.
PSG Aruna Sri and Anusha M., "Big Data-Survey,"
Indonesian Journal of Electrical Engineering and
Informatics (IJEEI), vol. 4, no. 1, pp. 74-80, MArch
2016, DOI: 10.11591/ijeei.v4i1.195.
Doug Laney. (2001, February)
M. Van Rijmenam. Why The 3V’s Are Not Sufficient To
Describe Big Data. Web.
Timothy Schultz, "Turning healthcare challenges into big
data opportunities: A use-case review across the
pharmaceutical development lifecycle," Bulletin of the
Association for Information Science and Technology,
June 2013.
Dr N M Saravan , Kumar, T. Eswari, P. Sampath, and S.
Lavanya, "Predictive Methodology for Diabetic Data
Analysis in Big Data ," Procedia Computer Science,
vol. 50, pp. 203-208, 2015.
Dan Ciregan, Ueli Meier, and Jürgen Schmidhuber, "Multi-
column deep neural networks for image classification,"
in Computer Vision and Pattern Recognition
(CVPR),IEEE Conference on Computer Vision and
Pattern Recognition, 2012.
Dileep Kumar Patel, Tanmoy Som, Sushil Kumar Yadav,
and Manoj Kumar Singh, "Handwritten Character
Recognition Using Multiresolution Technique and
Euclidean Distance Metric," Journal of Signal and
Information Processing, vol. 3, pp. 208-214, May 2012.
Roman Filipovych and Christos Davatzikos, "Semi-
supervised pattern classification of medical images:
Application to mild cognitive impairment (MCI),"
NeuroImage, vol. 55, no. 3, pp. 1109-1119, April 2001.
Sergei Kucheryavski, "Using hard and soft models for
classification of medical images," Chemometrics and
Intelligent Laboratory Systems, vol. 88, no. 1, pp. 100-
106, Auguest 2007.
Maria-Luiza Antonie, Osmar R. Zaiane and Alexadru
Coman, "Application of Data Mining Techniques for
Medical Image Classification," in Proceedings of the
Second International Conference on Multimedia Data
Mining in conjunction with ACM SIGIKDD
Conference, San Francisco, pp. 94-101, 2001.
Jeffrey Ullman and Anand Rajaraman, Mining of Massive
Datasets., 2011.
Julie Beth Lovins, "Development of a Stemming
Algorithm," Mechanical Translation and
Computational Linguistics, vol. 11, no. 1 and 2, March
and June 1968.
Isabelle Guyon and André Elisseeff, "An Introduction to
Feature Extraction," in Jouran of Machine Laerning
Research 3, 1157-1182, 2003.
B. D. Ripley, Pattern Recognition and Neural Networks.:
Cambridge University Press, 1996.
Mohamed Aly, "Survey on Multiclass Classification
Methods," Caltech, Technical 2005.
Cliff Click, Michal Malohlava, Arno Candel, Hank Roark,
and Viraj Parmar. (2017, April) Gradient Boosting
Machine with H2O.
Arno Candel, Viraj Parmar, Erin Ledell, and Anisha Arora.
(2015, March) Deep Learning with H2O.
H2O, (10 Jan. 2016) http://docs.h2o.ai/h2o/latest-stable/
h2o-docs/data-science/drf.html.
Andrew B. Collier. (2015) Making Sense of Logarithmic
Loss. Exegetic Analytics.
Northwestern University, Centre for Genetic Medicine, and
University of Maryland School of Medicine Institute
for Genome Sciences, doid-non-classified.obo, format-
version: 1.2; data-version: releases/2017-04-13.
Roger Henderson. (2016, April), http://www.netdoctor.co.
uk/health-services/nhs/a4502/a-to-z-of-hospital-
departments/
Identifying a Medical Department based on Unstructured Data
481

Mayoclinic, (10 Jan. 2016)http://www.mayoclinic.org/
departments-centers/index.
Jiawei Han and Micheline Kamber, Data Mining: Concepts
and Techniqes, 2nd ed.: Morgan Kaufmann Publishers,
2001.
B. L. Kalman and S. C. Kwasny, "Why tanh: choosing a
sigmoidal function," in [Proceedings 1992] IJCNN
International Joint Coference on Neural Networks, vol.
4, Baltimore, MD, 1992, pp. 578-581, doi:
10.1109/IJCNN.1992.227257.
R. Hahnloser, R. Sarpeshkar, M. A. Mahowald, R. J.
Douglas, and H.S. Seung, "Digital Selection and
analogue amplification coexist in a cortex-inspired
silicon circuit," Nature, vol. 405, pp. 947-951, 2000.
I.J. Goodfellow, D. Warde-Farley, M. Mirza, A. Courtville,
and Y. Bengio, "Maxout networks," in Proceedings of
the 30th International Conference on Machine
Learning, 2013, pp. 1319-1327.
Nitish Srivastava, Geoffrey Hinton, Alex Krizhevsky, Ilya
Sutskever, and Ruslan Salakhutdinov, "Dropout: A
Simple Way to Prevent Neural Networks from
Overfitting," Journal of Machine Learning Research,
vol. 15, pp. 1929-1958, 2014.
J. H. Friedman, "Greedy function approximation: a gradient
boosting machine," Statistics, Stanford Universiy,
Technical 1999.
J. H. Friedman, "Stochastic gradient boosting," Comput.
Stat. Data Anal., vol. 38, no. 4, pp. 367-378, 2002
ICEIS 2018 - 20th International Conference on Enterprise Information Systems
482
