
Phenotypic and Molecular Detection of OXA-48 Gene
Carbapenem-resistant Klebsiella Pneumoniae and Escherichia Coli
Isolates in Haji Adam Malik Hospital Medan, Indonesia
Mirzan Hasibuan
1,2
, R. Lia Kusumawati
2,3,4*
and Dwi Suryanto
3
1
Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Sumatera Utara, Jalan Bioteknologi
No. 1 Kampus USU, Medan 20155, Indonesia
2
University of Sumatera Utara Hospital, Jalan dr. T Mansyur No. 66 Kampus USU Medan 20154, Indonesia
3
Department of Microbiology, Faculty of Medicine, Universitas Sumatera Utara, Jalan Universitas No. 1 Kampus USU
Medan 20155, Indonesia
4
Haji Adam Malik Hospital Medan, Jl. Bunga Lau No.17, Kemenangan Tani, Medan Tuntungan, Kota Medan, Sumatera
Utara 20136, Indonesia
Keywords: Phenotypic, Molecular Detection, OXA-48, Carbapem-Resistant, Carbaenemase
Abstract: Carbapenem-resistant to Klebsiella pneumoniae and Escherichia coli, increasingly reported as a major cause
of infection in hospitals and healthcare facilities. Carbapenemase is an enzyme produced by gram-negative
bacteria that causes failure of antibiotic therapy, especially the carbapenem. The study aimed to characterize
phenotypically using Vitek 2 Compact and detect OXA-48 clinical isolates of Klebsiella pneumoniae and
Escherichia coli belonging to Carbapenem-Resistant using PCR. The results showed that all were beta-
lactamase producers, which of 12 (14.11%) were resistant to carbapenem. The phenotype distribution of
carbapenem is 10/12 (11.75%) Klebsiella pneumoniae and 2/12 (2.35%) Escherichia coli. From 12 isolates
Carbapenemase phenotypically, in which 10 (11.77%) of both bacteria bearing OXA-48 gene with the
distribution 9 (10.6%) Klebsiella pneumoniae and 1 (1.17%) Escherichia coli, respectively. The study
shows that prevalence of OXA-48 genes in North Sumatra, Indonesia in Klebsiella pneumoniae and
Escherichia coli which cause failure of therapeutic types of antibiotics Carbapenem. The only antibiotic that
is still sensitive to carbapenem-resistant bacteria based on antimicrobial susceptibility is amikacin, which
can be recommended as carbapenemases therapy.
1 INTRODUCTION
The spread of resistance to carbapenem by
Enterobacteriaceae, especially Klebsiella
pneumoniae and Escherichia coli (Maryam et.al,
2017). The emergence of bacterial resistance to the
carbapenem is a global problem that is growing
rapidly and requires urgent action for the
international scientific community (CDC, 2013).
Tropical countries such as Indonesia, infections by
Klebsiella pneumoniae and Escherichia coli are
strongly associated with health care such as the use
of health facilities that cause urinary tract infections,
as well as postoperative wound care (Raka et.al,
2006).
Carbapenem-resistant over the past decade,
healthcare settings have emerged due to the bacterial
infection of Enterobacteriaceae and are strongly
related to the ability of bacteria to produce β-
lactamase, which is capable of hydrolyzing
carbapenems (Nordmann et.al, 2012). Most of these
phenomena are related to the spread of various types
of β-lactamases. Carbapenem-hydrolysing β-
lactamase major in Enterobacteriaceae is class A
Klebsiella pneumoniae carbapenemases and class B
acquires Metallo beta-lactamases (MBLs) which is
dominated by Klebsiella pneumoniae which has
shown rapid international spread. In the D-class β-
lactamase OXA-48 despite its weak activity in
resistance but is increasingly reported to be
significant in Enterobacteriaceae. General
epidemiological observations and recent research
shows that OXA-48 producers are increasingly
being identified in various developing countries
(Nordmann et.al, 2014).
590
Hasibuan, M., Kusumawati, R. and Suryanto, D.
Phenotypic and Molecular Detection of OXA-48 Gene Carbapenem-resistant Klebsiella Pneumoniae and Escherichia Coli Isolates in Haji Adam Malik Hospital Medan, Indonesia.
DOI: 10.5220/0010079805900594
In Proceedings of the International Conference of Science, Technology, Engineering, Environmental and Ramification Researches (ICOSTEERR 2018) - Research in Industry 4.0, pages
590-594
ISBN: 978-989-758-449-7
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
The increasing prevalence of carbapenem-
resistant almost all over the world provides the basis
for the importance of phenotypic characterization
and molecular detection of the Carbapenemase gene
in the Laboratory of Clinical Microbiology.
Molecular detection of the OXA-48 gene in bacteria
that produce carbapenemase enzymes is needed to
control antibiotic resistance, avoid bacterial
transmission and improve therapy management. The
aim of the study was to characterize phenotypically
and detect OXA-48 genes in isolates of Klebsiella
pneumoniae and in Escherichia coli resistant to
carbapenem in Haji Adam Malik Hospital Medan,
Indonesia.
2 METHOD
2.1 Phenotypic Characterization
Phenotypic characterization was carried out by
identification of bacterial types and sensitivity
testing of antibiotics as well as carbapenemase
phenotype using Vitek 2 Compact. The sample was
taken purely sampling based on germ pattern in the
first semester of 2015. Bacterial isolates were
obtained from routine examination of the Laboratory
of Clinical Microbiology, Installation of Diagnostic
Laboratory Haji Adam Malik Hospital Medan.
2.2 Isolation of DNA
Isolation of DNA was isolated by the freeze-thaw
cycling method before the isolate was subcultured
on Mac-Conkey Agar medium with a 24-hour
incubation period at 37°C. One colony was put into
a tube containing 20μl of aquabidest and
homogenized, then frozen for 10 minutes at 20°C,
followed by heating for 10 minutes at 90°C, hot and
frozen cycles were carried out 6 cycles. The
isolation of DNA was centrifuged for 5 minutes at a
speed of 13.000 rpm. The supernatant part is
separated, DNA purity is measured using a
nanophotometer.
2.3 Molecular Detection of OXA-48
Molecular detection using specific primers:
GCG-TGG-TTA-AGG-ATG-AAC-AC forward and
CAT-CAA-GTT-CAA-CCC-AAC-CG reverse. PCR
preparation began with DNA amplification at 25μl,
consisting of 12.5μl mastermix green go-Taq, 8.5µl
nuclease-free water, 1μl of forward and reverse
primer and 2μl bacterial DNA. Amplification is
carried out on Thermocycling reactions. PCR results
were electrophoresed and documented in UV
Reader.
3 RESULTS AND DISCUSSIONS
3.1 Phenotype
Profile of the Vitek 2 Compact antimicrobial
susceptibility from 85 clinical isolates showed that
all isolates are ESBLs, consisting of 52% of
Klebsiella pneumoniae and 48% of Escherichia coli,
respectively. The percentage profile of antimicrobial
sensitivity of the samples is presented in Table 1.
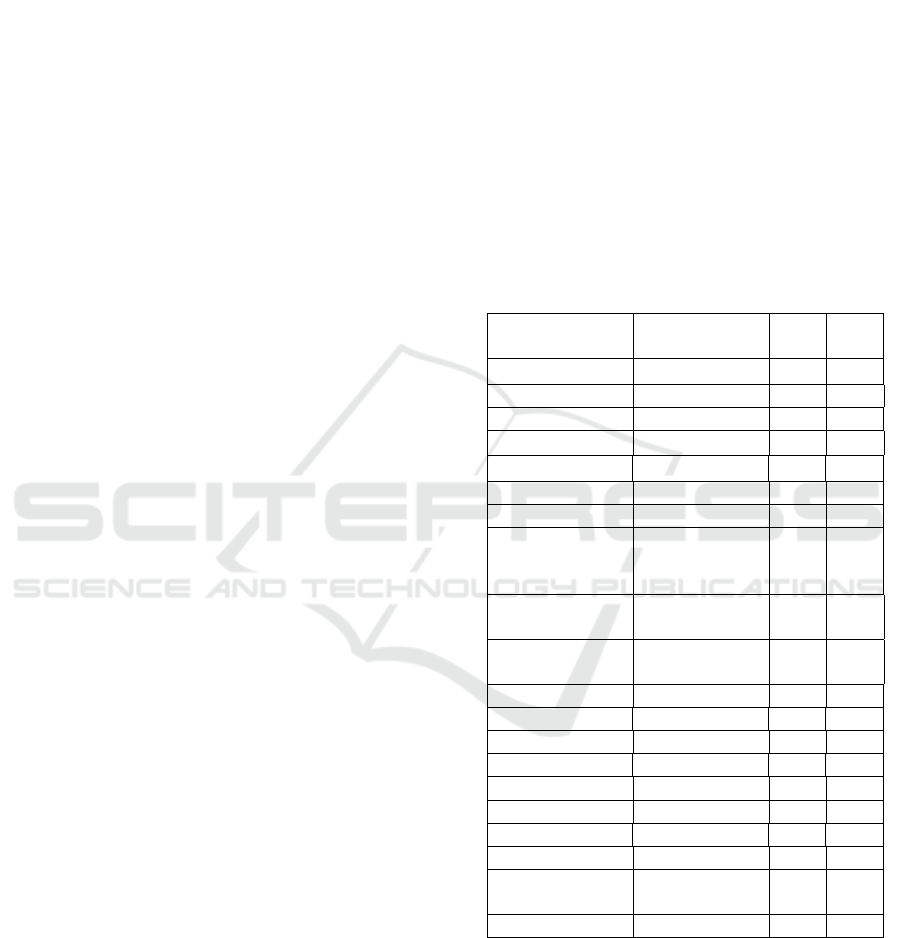
Table 1: Percentage of Antibiotic Sensitivity Tests.
Group Antibiotics
Agents
S
(%)
R
(%)
Penicillin Amoxycillin 0 100
Ampicillin 0 100
Cephalosporin Cefotaxime 0 100
Ceftriaxone 0 100
Ceftazidime 0 100
Cefepime 0 100
Monobactam Aztereonam 0 100
Beta-lactamase
Inhibitor
Amoxicillin/
Clavulanic
Acid
27 73.0
Piperacillin/
Tazobactam
43.5 56.4
Cefoperazone/
Sulbactam
63.5 36.4
Aminoglicosides Gentamycin 35.3 64.7
Amikacin 100 0
Carbapenem Ertapenem 85.9 14.1
Meropenem 85.9 14.1
Imipenem 85.9 14.1
Fluoroquinolone Ciprofloxacin 0 100
Levofloxacin 25.8 74.2
Fosfomycin Fosfomycin 86.5 13.5
Folate Pathway
Inhibitor
Cotrimoxazole 0 100
Tigecil Tigecycline 84.5 15.5
All isolates showed resistance (R) to beta-lactam
group antibiotics (penicillins, monobactams,
cephalosporins), which indicates that all isolates
carrying Extended-Spectrum Beta-Lactamases
(ESBLs). Resistance was also shown in the beta-
lactamase inhibitor group of 73% amoxicillin/
clavulanic acid, 56% piperacillin/tazobactam and
Phenotypic and Molecular Detection of OXA-48 Gene Carbapenem-resistant Klebsiella Pneumoniae and Escherichia Coli Isolates in Haji
Adam Malik Hospital Medan, Indonesia
591
36.4% cefoperazone/sulbactam, respectively.
Resistance is also shown in other classes of
antibiotics such as ciprofloxacin and cotrimoxazole.
From 85 isolate were found to be 12 (14.2%)
resistant to carbapenem (ertapenem, meropenem
imipenem), of which 2 (2.35%) Escherichia coli and
10 (11.77%) Klebsiella pneumoniae. which
indicated that the isolates are carrying ESBls and
Carbapenemases. The only antibiotic that is still
sensitive (S) to all isolates based on antimicrobial
susceptibility is amikacin.
Resistance occurs because of the genes that
encode resistant and expressed phenotypically.
Resistance occurs because of the genes that encode
resistant and expressed phenotypically. This is very
important because enzymes produced by bacteria
will express genes for resistance (Thenmozhi et.al,
2014). The reliability of the Vitek 2 Compact
System as an ESBLs and Carbapenemases detection
system was verified in comparison with another
method such as double disc synergy which is
recommended by Clinical Laboratory Standards
Institute. The characterization of the Carbapenemase
phenotype with a combination of Modified Hodge
Test (MHT) using discs and ertapenem and
meropenem is preferred. In addition, the MBL strip
E-test was also used to evaluate Metallo-β-lactamase
production (CLSI, 2013).
From the results of the Carbapenemase
phenotyping using Vitek 2 Compact, it is necessary
to proceed at the molecular level to detect ESBLs
encoding genes as well as the accuracy of both
methods. Then amplification and molecular
detection of OXA-48 gene against these 12 isolates
by using PCR.
3.2
Molecular Detection of OXA-48
Gene
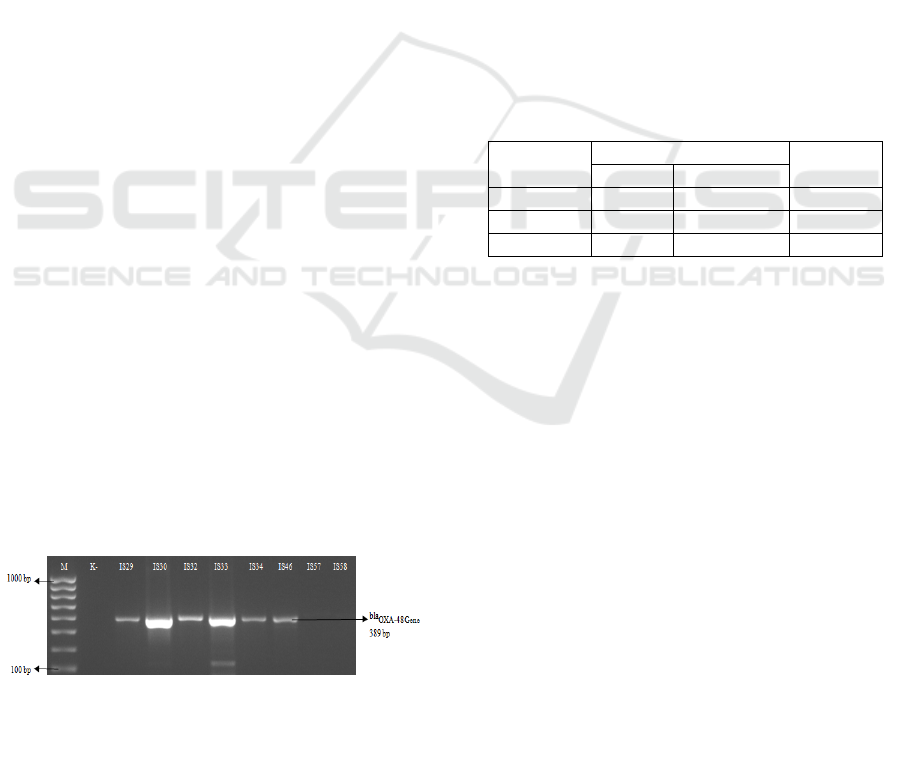
Molecular detection of the OXA-48 gene showed
DNA amplification 389 bp in Klebsiella pneumoniae
and Escherichia coli (figure 1.). Based in this study
found 10/12 OXA-48 gene with the percentage of
11.77%.
Figure 1: profiles of OXA-48 gene
K- (Escherichia coli Non-Carbapenemase),
IS29 (Klebsiella pneumoniae 11474),
IS30 (Klebsiella pneumoniae 11527),
IS32 (Klebsiella pneumoniae 11583),
IS33 (Klebsiella pneumoniae 11665),
IS34 (Klebsiella pneumoniae 11417),
IS46 (Klebsiella pneumoniae 11311),
IS57 (Escherichia coli 11926),
IS58 (Klebsiella pneumoniae 11688).
From Figure 1 it can be seen that the appearance of
the DNA band pattern indicates that the bacteria has
the
OXA-48 gene. Proving that the primary can be
used OXA-48 produces 389 bp amplicon. In the
previous study, detecting the OXA-48 gene using
the same primary, where 27 of the 28 isolates of the
Motahari Hospital, Tehran, Iran (Azimi et.al, 2014).
From 12 isolates carbapenemase phenotypically,
it was found that 10 (11.77%) Klebsiella
pneumoniae and 2 (2.35%) of Escherichia coli
isolates showed carbapenemase, isolates containing
OXA-48 gene were 10 (11.77%) which are
presented in table 2. Another 2.35% possibility
contains other genes that cause carbapenem-
resistance.
Table 2: Percentage of ESBLs, Carbapenemases
phenotype and OXA-48 gene.
Bacterial
species
Phenotype OXA-48
gene
ESBLs Carbapenemase
K.pneumoniae 44 (52%) 10(11.77%) 9(10.6%)
E.coli 41 (48%) 2(2.35%) 1(1.17%)
total : 85(100%) 12 (14.12%) 10(11.77%)
The results of this study showed the spread of OXA-
48 gene in Indonesia. In this study, the OXA-48
gene was not only found in Klebsiella pneumoniae,
but was also found in the Escherichia coli. In 2
phenotypic Carbapenemase isolates, not found the
blaOXA-48 gene. There may be other genes such as
IMP-1, NDM-1 and other types of carbapenemase
genes. Molecular detect of ESBLs genes by
Karuniawati et.al (2013) in RSPN Cipto
Mangunkusumo Jakarta, Indonesia. From the 61
Gram-negative bacteria-producing carbapenemases
in phenotype, did not have the OXA-48 type gene.
However, the gene encoded carbapenemases such as
IMP-1 and NDM-1.
The presentation of carbapenem-resistant in
Enterobacteriaceae poses a major problem for health
services, for example the limited choice of antibiotic
therapy. As a bacterium producing Carbapenemase,
these bacteria are not only resistant to carbapenem
but almost resistant to all beta-lactam groups, except
monobactam (aztreonam) for MBL and other
compounds such as OXA-48 (Jacoby et.al, 2004). In
this study, it was found that all isolates were even
resistant to aztreonam. In addition, the extended
spectrum β-lactamase (ESBL) is strongly associated
ICOSTEERR 2018 - International Conference of Science, Technology, Engineering, Environmental and Ramification Researches
592

with carbapenem resistance in Enterobacteriaceae,
this is due to the female bacteria producing AmpC β-
lactamase or loss of porin in the bacterial cell wall
(Woodford, 2007).
This gene OXA-48 is also known as the
carbapenemase coding gene in gram-negative
bacteria. The first identified OXA-48 gene
manufacturer was derived from the Klebsiella
pneumoniae strain isolated in Turkey in 2003. Since
then, manufacturers of
bla
OXA-48 have been widely
reported in Turkey as a source of bacteria that cause
nosocomial infections (Nordmann et.al, 2004). The
worldwide distribution of OXA-48 manufacturers
now covers countries in Europe, Africa, America,
and even Asia. The spread of multi-resistant
pathogens worldwide has been linked to a variety of
epidemiological factors including international
patient transfer originating from endemic areas
(Girmenia et.al, 2016). The argument about the
spread of Klebsiella pneumoniae as a type of
bacteria that causes carbapenemase is considered a
high-risk organism (Munoz-Price et.al, 2013).
The threat of antimicrobial resistance has been
recognized by World Health Organization, where
this threat involves and requires all actions related to
the agency and society as a whole (WHO, 2013).
Clone expansion is a major driver of the spread of
carbapenemase in gram-negative bacteria, especially
in Enterobacteriaceae. In addition, carbapenemase
transmission as a clonal lineage of
Enterobacteriaceae (CPE) is stable in the defense of
the carbapenemase coding gene. Horizontal transfer
of this type of gene is very likely to occur through
the moving genetic element, the plasmid (Kitchel
et.al, 2009).
Study on the detection of OXA-48 genes is still
rare in Indonesia, especially in North Sumatera. This
study shows the presence of the OXA-48 gene as a
cause of Carbapenemase in Klebsiella penumoniae
and Escherichia coli. This study also provides an
overview of the prevalence and incidence of
resistance to carbapenem, for this reason the
awareness of all hospitals is the importance of
controlling resistance in the present and avoiding the
transmission of resistant bacteria in health facilities.
4 CONCLUSIONS
The prevalence of carbapenemase has been shown in
this study, found of the phenotype 12 (14.12%) and
molecularly detected 10 (11.77%) OXA-48 genes.
However, there were 2 other isolates that were not
found in the OXA gene, possibly another gene that
causes carbapenemase.
ACKNOWLEDGEMENTS
The author would to thanks Director of Haji Adam
Malik General Hospital Medan, Indonesia,
especially for Clinical Microbiology Laboratory.
REFERENCES
Maryam A, Ali A, Manal A and Hisham A. 2017.
Comparison of phenotypic and PCR methods for
detection of carbapenemases production by
Enterobacteriaceae. Saudi Journal of Biological
Sciences. ELSEVIER.
Centers for Disease Control and Prevention. 2013. Vital
signs: carbapenem-resistant
Enterobacteriaceae. MMWR Morbid. Mortal. Wkly.
Rep. CDC.
Raka, LD, Zoutman G, Mulliqi S, Krasniqi I, Dedushaj N.
2006. Prevalence of nosocomial infections in high-risk
units in the university clinical center of Kosova.
Infection Control Hospital Epidemiology. SHEA.
Nordmann P, Dortet L, and Poirel L. 2012. Carbapenem
resistance in Enterobacteriaceae: here is the storm!.
Trends in Molecular Medicine. CE Press.
Nordmann, P and Poirel, L. 2014. The difficult-to-control
spread of carbapenemase producers in
Enterobacteriaceaeworldwide. Clin Microbio Infect.
ELSEVIER.
Azimi A, Nordmann P and Bonin R. 2014. First report of
OXA-48-producing Klebsiella pneumoniae strains in
Iran. GMS Hygiene and Infection Control. PMC.
Karuniawati A, Rosa Y, Saharman, Delly C and Lestari.
2013. Detection of Carbapenemase Encoding Genes in
Enterobacteriace, Pseudomonas aeruginosa, and
Acinetobacter baumannii Isolated from Patients at
Intensive Care Unit Cipto Mangunkusumo Hospital in
2011. The Indonesian Journal of Internal Medicine.
Acta Medica Indonesiana.
Jacoby GA, Mills DM, Chow N. 2004. Role of beta-
lactamases and porins in resistance to ertapenem and
other beta-lactams in Klebsiella
pneumoniae. Antimicrob Agents Chemother. American
Society for Microbiology.
Woodford N. 2007. Ertapenem resistance among
Klebsiella and Enterobacter submitted in the UK to a
reference laboratory. Int J Antimicrob Agents.
American Society for Microbiology.
Nordmann P, Naas T and Poirel L. 2004. Global Spread of
Carbapenemase Producing Enterobacteriaceae.
Emergency Infection Disease. PMC.
Girmenia C, Serrao A and Chanichella M. 2016.
Epidemiology of Carbapenem Resistant Klebsiella
pneumoniae Infections in Mediterranean Countries.
Journal of Hematology and Infectious Diseases.
MJHID.
Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ,
Daikos GL, Cormican M, Cornaglia G, Garau J,
Gniadkowski M, Hayden MK. 2013. Clinical
Phenotypic and Molecular Detection of OXA-48 Gene Carbapenem-resistant Klebsiella Pneumoniae and Escherichia Coli Isolates in Haji
Adam Malik Hospital Medan, Indonesia
593

epidemiology of the global expansion of Klebsiella
pneumoniae carbapenemases. Lancet Infect Dis. The
Lancet.
World Health Organization. 2014. Antimicrobial
Resistance (AMR): Global Report on Surveillance.
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-
Venezia S, Carmeli Y, Brolund A, Giske CG. 2009.
Molecular epidemiology of KPC-producing Klebsiella
pneumoniae isolates in the United States: clonal
expansion of multilocus sequence type
258. Antimicrob Agents Chemother. AAC.
Thenmozhi S, Moorthyl K, Suresh T, Suresh M. 2014.
Antibiotic Resistance Mechanism of ESBL Producing
Enterobacteriaceae in Clinical Field: A Review. Int. J.
Pure App. Biosci. ELSEVIER.
Hackman H, Adje G, Gordon A, Laryea E , Quay S. 2013.
The Reliability of Using Vitek 2 Compact System to
Detect Extended-Spectrum Beta-lactamase-producing
Isolates in Escherichia coli and Klebsiella pneumoniae
in Accra, Ghana. J IISTE Life Science and
Technology. IISTE.
Performance Standards for antimicrobial susceptibility
testing, 23rd informational supplement. CLSI
document M100-S23. Clinical and Laboratory
Standards Institute. 2013.
ICOSTEERR 2018 - International Conference of Science, Technology, Engineering, Environmental and Ramification Researches
594
