
Entorhinal Grid Cells May Facilitate Pattern Separation
in the Hippocampus
Jochen Kerdels and Gabriele Peters
University of Hagen, Universit
¨
atsstrasse 1, D-58097 Hagen, Germany
Keywords:
Pattern Separation, Grid Cells, Entorhinal Cortex, Dentate Gyrus, Hippocampus.
Abstract:
The dentate gyrus (DG) in the hippocampus of the mammalian brain is known to exhibit strong pattern sepa-
ration. However, how this pattern separation arises in the DG is not well understood. Here we offer a novel
hypothesis regarding this problem by demonstrating that pattern separation can already be performed by en-
torhinal grid cells, which are located just one synapse upstream of the DG. For our simulations we utilize a
recently introduced grid cell model that interprets the behavior of grid cells as just one instance of a general
information processing scheme. The obtained results challenge the established view that pattern separation
occurs primarily in the DG, and they uncover a common misconception regarding the specificity of ensemble
activity in grid cells.
1 INTRODUCTION
The parahippocampal region (PHR) and the hip-
pocampal formation (HF) in the mammalian brain
are vital for storing and retrieving episodic memories,
i.e., memories of specific events experienced by the
organism (Tulving and Markowitsch, 1998; Burgess
et al., 2002; Rolls, 2013). A key requirement for this
kind of memory is the ability to distinguish between
similar events that may differ only in minute details.
To accomplish such differentiation it is commonly as-
sumed that parts of the hippocampus carry out some
form of pattern separation that orthogonalizes the in-
put signals before they are stored in an autoassociative
memory (Leutgeb et al., 2007; Yassa and Stark, 2011;
Rolls, 2013). As a result of this pattern separation,
similar input signals cause divergent sets of neurons
to become active and thus reduce confusion of these
inputs in subsequent processing steps. Within the hip-
pocampus such divergent activation of neurons in re-
sponse to similar inputs was primarily observed in
the dentate gyrus (DG) (Leutgeb et al., 2007; Berron
et al., 2016). However, how this observed pattern sep-
aration in the DG arises is not well understood (Rolls,
2013).
In this paper we offer a novel hypothesis regarding
this problem. Based on our recently introduced com-
putational model of entorhinal grid cells (Kerdels and
Peters, 2015; Kerdels, 2016) we will show that the be-
havior of grid cells – if it is interpreted as an instance
of a general information processing scheme – already
results in strong pattern separation within the entorhi-
nal cortex (EC). As the latter provides the main input
to the dentate gyrus (van Strien et al., 2009) we hy-
pothesize that the pattern separation observed in DG
is facilitated by output signals from the EC that are
already well separated and that the DG itself just im-
proves this separation by sparsification.
The next two sections provide a brief overview
of the parahippocampal-hippocampal network (PHR-
HF) and our computational model of entorhinal grid
cells. Section 4 outlines the experimental setup and
characterizes the input signal used in the simulations.
In sections 5 and 6 we present and discuss the simu-
lation results.
2 PHR-HF OVERVIEW
The parahippocampal-hippocampal region of the
mammalian brain is part of the limbic system. It
has a long evolutionary history and putative homo-
logues can be found in most vertebrate species (But-
ler and Hodos, 2005). Functionally, the region is nec-
essary for short term and declarative memory, which
includes factual knowledge as well as episodic mem-
ories (Squire et al., 2008).
The parahippocampal region (PHR) consists of
five main areas designated as perirhinal cortex (PER),
postrhinal cortex (POR), presubiculum (PrS), para-
Kerdels J. and Peters G.
Entorhinal Grid Cells May Facilitate Pattern Separation in the Hippocampus.
DOI: 10.5220/0006514601410148
In Proceedings of the 9th International Joint Conference on Computational Intelligence (IJCCI 2017), pages 141-148
ISBN: 978-989-758-274-5
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
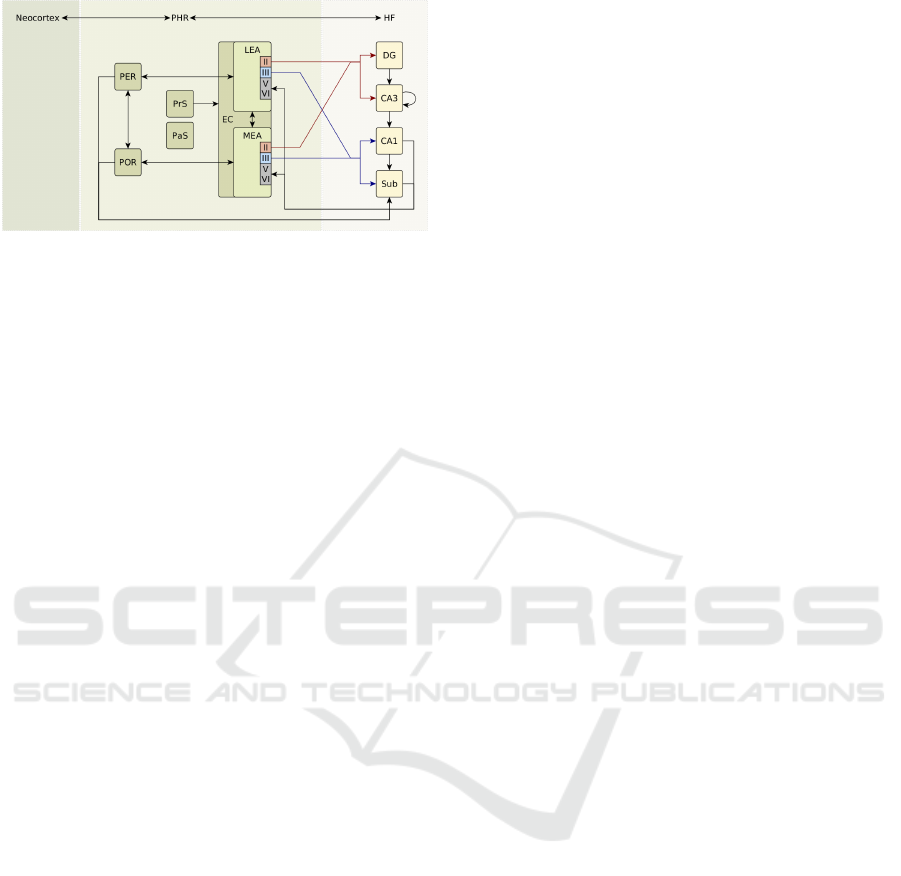
Figure 1: The standard view of the parahippocampal-
hippocampal network based on the review by (van Strien
et al., 2009).
subiculum (PaS), and entorhinal cortex (EC). The lat-
ter being further subdivided into a lateral (LEA) and
a medial (MEA) part. The areas of the PHR have six
layers similar to the neocortex with layers II/III and
layers V/VI being the principal cell layers that contain
the majority of neuronal cell bodies. The hippocam-
pal formation (HF) consists of four main areas desig-
nated as dentate gyrus (DG), cornu ammonis 3 (CA3),
cornu ammonis 1 (CA1), and subiculum (Sub). In
contrast to the PHR and the neocortex the areas of the
HF have only three layers, with the central layer con-
taining the majority of cell bodies (Witter et al., 2000;
Squire et al., 2008; van Strien et al., 2009; Somogyi,
2010).
Figure 1 outlines the basic PHR-HF network. It
shows that the PHR acts as a gateway that mediates
both the input from the neocortex to the HF, as well
as the output from the HF to the neocortex. Two ma-
jor, bidirectional projection streams facilitate this ex-
change of signals. The PER connects predominantly
with the LEA, while the POR connects to the MEA.
From the upper layers II/III of the EC the so called
perforant pathway projects then to all areas of the HF.
Signals from the second layer reach the DG and CA1,
whereas signals from the third layer go to CA1 and the
subiculum. The output from the HF projects from ar-
eas CA1 and the subiculum to deep layers V/VI of the
EC (van Strien et al., 2009). As a rough approxima-
tion the hippocampus can be interpreted as a sequence
of processing modules, starting with the DG and end-
ing with the subiculum, that receives inputs from the
upper layers of the EC and feeds its results back to
the lower layers of the EC. As such, the EC can be
interpreted as the main interface to the hippocampus.
A major advance in the understanding of EC func-
tion resulted from the discovery of grid cells and their
subsequent investigation (Fyhn et al., 2004; Hafting
et al., 2005; Rowland et al., 2016). The activity of
grid cells correlates strongly with the animal’s loca-
tion creating a hexagonal pattern of firing fields that
spans the entire environment of the animal. This
strong spatial correlation provides a rare opportunity
to experimentally observe and interpret the behav-
ior of neurons in this part of the brain. Common
hypothesis regarding the function of grid cells view
these cells as specialized components in a system fa-
cilitating orientation and navigation (Rowland et al.,
2016). However, recent observations of grid like fir-
ing patterns in contexts other than locomotion (Kil-
lian et al., 2012; Constantinescu et al., 2016; Aronov
et al., 2017; Diehl et al., 2017) may indicate that the
behavior of grid cells reflects a more general, underly-
ing processing scheme. To investigate this hypothesis
we developed a computational model of grid cells that
implements such a general processing scheme allow-
ing us to apply our model outside of the typical con-
text of navigation and orientation (Kerdels and Peters,
2015; Kerdels, 2016; Kerdels and Peters, 2016). We
briefly outline the model in the next section.
3 GRID CELL MODEL
We developed our grid cell model based on the idea
that the behavior observed in grid cells is just one
instance of a more general information processing
scheme. We hypothesize that grid cells form a simple,
piecewise representation of their entire input space by
learning a limited number of input patterns or proto-
types that reflect the input space structure. Simultane-
ously, competition among cells within a local grid cell
group ensures that the simple representations learned
by the individual cells are pairwise distinct and inter-
leave in such a way that a complex representation of
the input space emerges that is distributed over the
entire group of neurons.
On the neurobiological level such a prototype-
based representation could be stored within the den-
dritic tree of a neuron with individual prototypes
being stored in local dendritic subsections. In our
model we utilize the core ideas of the growing neural
gas (GNG) algorithm to describe this learning pro-
cess. The GNG is an unsupervised learning algorithm
that uses biologically plausible competitive Hebbian
learning to approximate the input space structure with
a network of prototypes (Martinetz and Schulten,
1994; Fritzke, 1995). We extended the regular GNG
algorithm into a recursive version (RGNG) that al-
lows us to simultaneously describe both the learning
processes of individual neurons as well as the com-
petition among a group of neurons by the same GNG
dynamics. For a formal description and an in-depth
characterization of the model we refer to (Kerdels and
Peters, 2016; Kerdels, 2016).
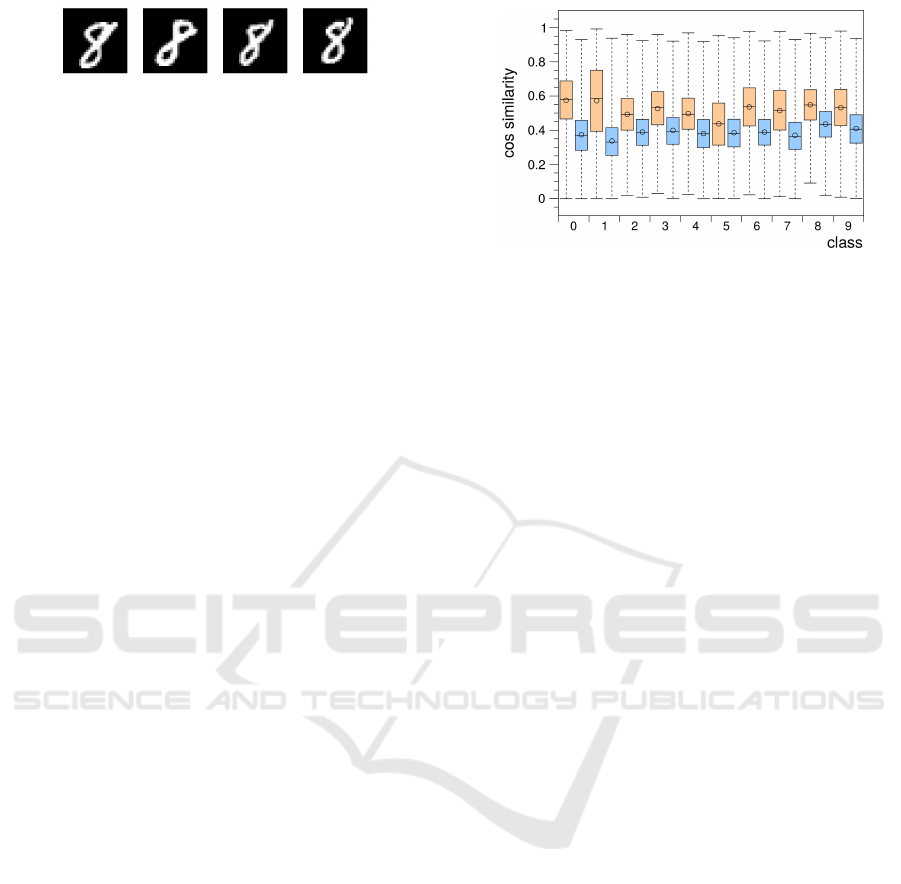
Figure 2: Typical samples of the digit 8 in the MNIST
database of handwritten digits (Lecun et al., 1998).
Regarding this work, the most important property of
the RGNG-based grid cell model is its ability to oper-
ate on arbitrary input spaces. Regardless of the par-
ticular input space, e.g., the modality or dimension of
the input signals, the modeled group of neurons will
try to learn the structure of that input space as well
as possible. In addition, as each neuron tries to learn
the structure of the entire input space the learned rep-
resentations of different neurons will be self-similar
and no individual neuron will “specialize” on a par-
ticular region of input space. As a consequence, small
shifts of the input signal are likely to cause a signif-
icant change in the set of active cells in the modeled
neuron group, hence effectively implementing a form
of pattern separation.
4 EXPERIMENTAL SETUP
To investigate the potential pattern separation capa-
bilities of grid cells we simulated multiple groups of
grid cells using the RGNG-based grid cell model and
exposed these groups to a sequence of input samples
from a given input space. For each input sample the
resulting activity of each simulated grid cell was de-
termined and the individual activities within a grid
cell group were then summarized in an activity vec-
tor. This vector can be interpreted as the grid cell
group’s encoding of the respective input sample. To
compare this encoding with the original encoding of
the input signal in terms of orthogonality we decided
to use the cosine similarity measure, as it is a direct
measure of the (non-)orthogonality of two vectors.
As input space we chose the well-known MNIST
database of handwritten digits (Lecun et al., 1998),
which provides a total of 60000 samples of hand-
written digits in the resolution of 28 × 28 = 784 pix-
els. Since the intra-class samples in this database ex-
hibit a high degree of similarity (Fig. 2) it is espe-
cially suited to investigate the pattern separation ca-
pabilities of an encoding scheme. Figure 3 shows the
intra- and inter-class cosine similarity distributions in
the MNIST dataset. Cosine similarity values close to
0 indicate orthogonal vectors, while values close to
1 indicate a high degree of similarity. For any en-
coding that performs pattern separation it is expected
that corresponding distributions of the encoded sig-
Figure 3: Box plot of the intra-class (orange, left columns)
and inter-class (blue, right columns) cosine similarity distri-
butions occurring in the MNIST dataset. Bottom and top of
dashed lines represent minimum and maximum values, bot-
tom and top of each box represent lower and upper quartiles,
thick lines represent medians, and circles represent mean
values of the distributions.
nals will be skewed towards 0.
A total of 15 grid cell groups were simulated. Each
group consisted of 100 neurons and received 2.88 mil-
lion (partial) input samples from the MNIST input
space, i.e., 48 repetitions of 60000 samples. Thus, the
activity vectors derived from each group were 100-
dimensional. The grid cell groups differed by the
number
{
20, 40, 80
}
of prototypes each neuron was
allowed to use for its input space representation, and
by processing either entire input samples or just the
top, bottom, left, or right half of the samples. The
number of prototypes per cell influences the “reso-
lution” with which the cell can form an input space
representation. A lower number of prototypes results
in a more coarse representation, i.e., grid cells with
a low number of prototypes have larger grid spacing
than grid cells with a high number of prototypes. The
numbers of prototypes used here were chosen from
a range that is biologically plausible (Kerdels, 2016).
The processing of partial input samples was used as
a proxy for simulating multimodal input, i.e., as input
that usually occurs together but that is represented and
encoded by independent groups of neurons. All other
parameters of the RGNG-based grid cell model were
held constant and are provided in the appendix.
5 RESULTS
Figure 4 shows the intra- and inter-class cosine sim-
ilarity distributions occurring in the activity vectors
of grid cell groups that processed 2.88 million in-
put samples from the MNIST input space and used
either 20, 40, or 80 prototypes per cell for their in-
put space representation. The activity vectors were
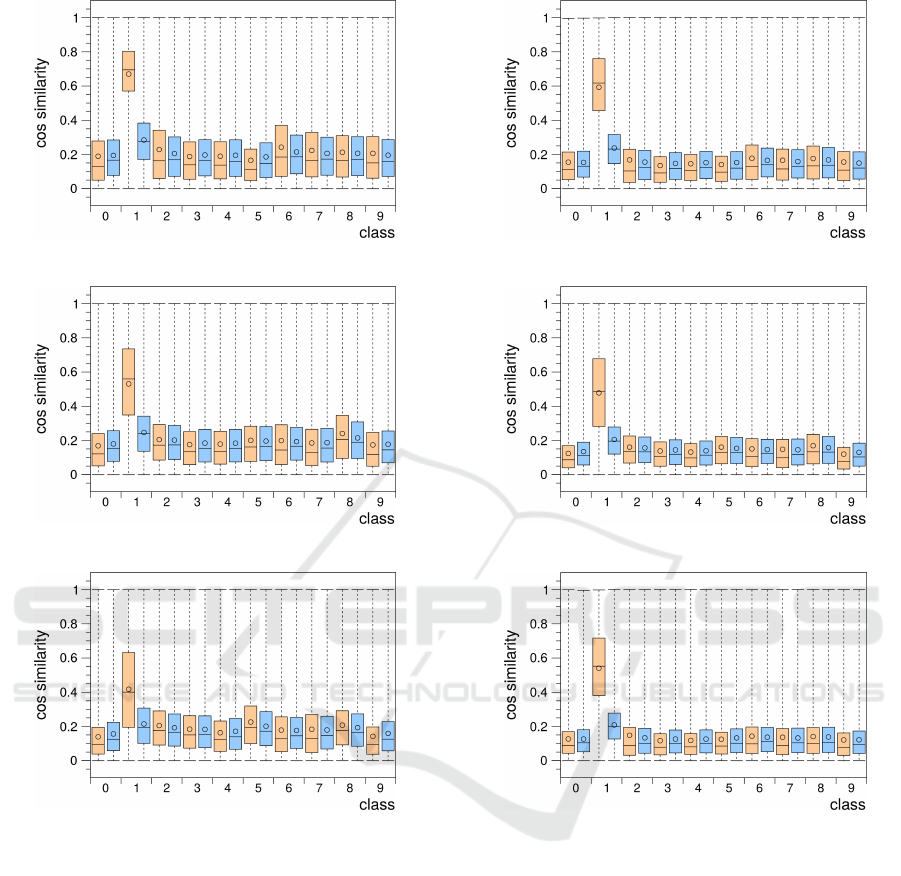
(a) 20 prototypes per cell
(b) 40 prototypes per cell
(c) 80 prototypes per cell
Figure 4: Box plots as in figure 3 of the intra- and inter-
class cosine similarity distributions occurring in the activity
vectors of grid cell groups using 20 (a), 40 (b), or 80 (c)
prototypes per cell for their input space representation.
sampled during the 48th input repetition of the 60000
MNIST training samples. For most classes the dis-
tributions show a significant decrease in the cosine
similarities when compared with the cosine similar-
ities measured in the MNIST set (Fig. 3). Mean val-
ues for both intra- and inter-class similarities dropped
from about 0.5 and 0.4, respectively, to about 0.2. In
addition, the clear difference of similarity values be-
tween intra- and inter-class distributions visible in the
MNIST set was equalized in the grid cell group activ-
ity vectors. Notably, the intra-class distribution of
(a) combined groups 20 and 40 prot.
(b) combined groups 40 and 80 prot.
(c) combined groups 20, 40, and 80 prot.
Figure 5: Box plots as in figure 3 of the intra- and inter-class
cosine similarity distributions occurring in combined activ-
ity vectors of grid cell groups. (a) Two grid cell groups us-
ing 20 and 40 prototypes per cell. (b) Two grid cell groups
using 40 and 80 prototypes per cell. (c) Three grid cell
groups using 20, 40, and 80 prototypes per cell.
class 1 deviates from these observations. It is broader
and retains a relatively high median similarity value.
In the case of the grid cell group that only uses 20 pro-
totypes per cell the median value has even increased
rather than decreased when compared to the MNIST
set median value of class 1. These deviations of the
class 1 distributions can be attributed to the fact that
the digit one is typically drawn as just a vertical stroke
with little variation.
It is a common notion within the grid cell literature
that the output signals of entorhinal grid cell groups
that exhibit different grid spacings combine in the hip-
pocampus to uniquely encode specific places in an
animal’s environment (Solstad et al., 2006). To in-
vestigate the orthogonality of such combined output
signals we concatenated the activity vectors of two
or more simulated grid cell groups that use differ-
ent numbers of prototypes per cell and analysed the
resulting intra- and inter-class cosine similarity dis-
tributions. Figure 5 shows the distributions occur-
ring in the combined activity vectors of groups us-
ing 20 and 40 prototypes (Fig. 5a), 40 and 80 proto-
types (Fig. 5b), as well as 20, 40, and 80 prototypes
(Fig. 5c). In general, the results are qualitatively simi-
lar to that from the non-combined, individual grid cell
groups (Fig. 4), though the widths of the distributions
as well as the mean similarity values have slightly de-
creased.
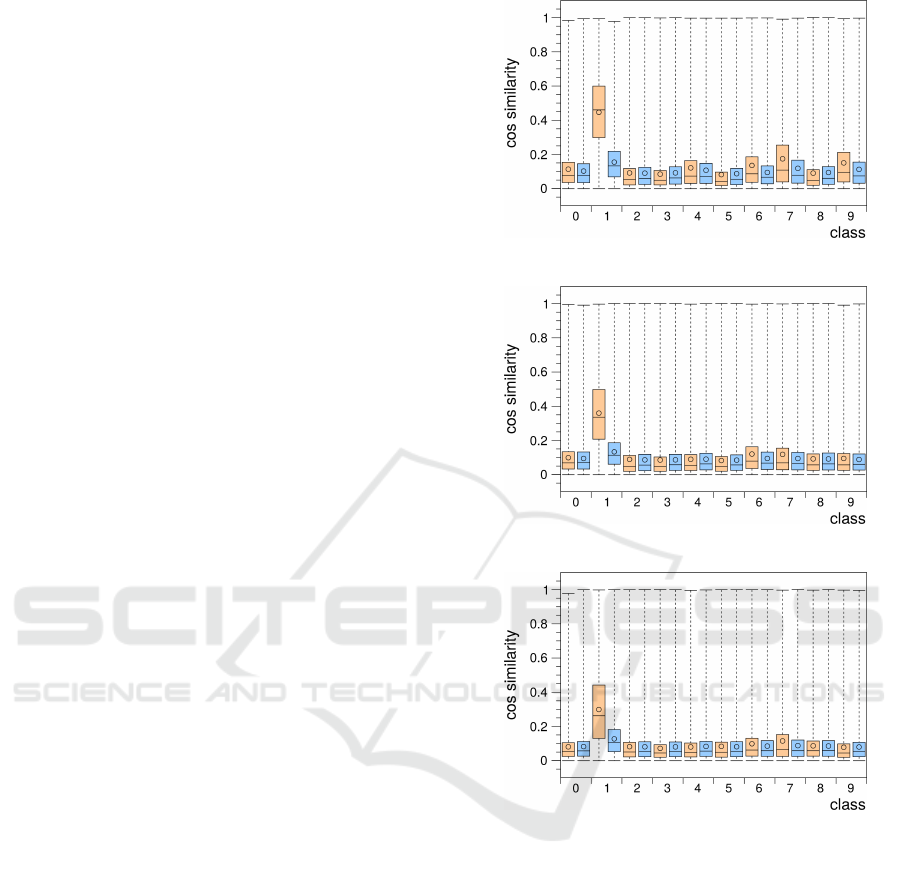
A different source of combined output signals can
be multimodal input. The entorhinal cortex receives
projections from various areas of the neocortex and
the limbic system. This multimodal input is then
processed and forwarded to the hippocampus. To
simulate input from multiple modalities we split the
MNIST input samples into top, bottom, left, and right
halves and let these partial samples be processed by
four separate grid cell groups. The activity vectors of
these four groups were then concatenated and anal-
ysed. Figure 6 shows the intra- and inter-class cosine
similarity distributions occurring in combined activity
vectors of groups that used 20 (Fig. 6a), 40 (Fig. 6b),
and 80 (Fig. 6c) prototypes per cell. In all three cases
there is a much more pronounced reduction in the
widths of the distributions and the mean similarity
values as compared to the distributions occurring in
the combined activity vectors of grid cell groups with
different grid spacings (Fig. 5). The lowest mean sim-
ilarity values of about 0.05 are achieved by the four
grid cell groups that use 80 prototypes per cell for
their input space representation. Remarkably, even
the mean similarity value of the intra-class distribu-
tion of class 1 decreases in this multimodal setting
down to a value of about 0.25 from about 0.6 mea-
sured in the MNIST set.
6 DISCUSSION
The results presented in the previous section indicate
that – under the given hypothesis – entorhinal grid
cells perform pattern separation on their input signals
and provide already orthogonalized output signals to
the hippocampus. This result provides a novel per-
(a) multi modal / 20 prototypes
(b) multi modal / 40 prototypes
(c) multi modal / 80 prototypes
Figure 6: Box plots as in figure 3 of the intra- and inter-
class cosine similarity distributions occurring in simulated
multimodal activity vectors of grid cell groups encoding
the top, bottom, left, or right halves of the MNIST input
samples using 20 (a), 40 (b), or 80 (c) prototypes per cell.
spective on the possible genesis of the pattern separa-
tion that can be observed in the dentate gyrus (Leut-
geb et al., 2007; Berron et al., 2016). From this per-
spective, the input signals to the DG may already be
well separated and the pattern separation observed in
the DG may just be a result of further sparsification,
e.g., due to the neurogenesis occurring in this region.
Furthermore, shifting the “functionality” of pattern
separation from DG to EC would provide orthogonal-
ized inputs not only to the DG but also to all other
parts of the hippocampus (Fig. 1). This shift would
improve the support of hippocampal memory models
that assume that memory retrieval can operate with-
out the involvement of the DG (Tulving and Markow-
itsch, 1998).
Another important aspect is the observation that
pattern separation does not require the collaboration
of multiple grid cell groups (Fig. 4). A typical grid
cell fires if the animal is at one of multiple locations.
In terms of the RGNG-based grid cell model the grid
cell fires if the input to the cell matches one of its
multiple prototypes. Thus, from the activity of a sin-
gle grid cell it is not possible to derive which of the
prototypes caused the cell to fire. The input pattern
could be similar to any of the multiple patterns en-
coded in the prototypes (Fig. 7). Within a grid cell
group the prototypes of grid cells are typically shifted
against each other. In most grid cell models found in
the literature this property is described in an idealized
way such that the relative locations of firing fields,
i.e., the prototypes of all grid cells in the group are
precisely aligned. In this idealized case the joint ac-
tivity of the grid cell group remains ambiguous with
respect to the actual input pattern. However, in real-
ity the alignment of firing patterns in a grid cell group
is less precise and can exhibit local permutations of
the cell’s firing fields. As a consequence, these per-
mutations make the joint activity of a grid cell group
much more specific and less ambiguous than is com-
monly thought. The RGNG-based grid cell model al-
lows for such natural variation in the alignment, and
the resulting specificity of the grid cell group activity
is reflected by the high degree of pattern separation
present in the inter-class distributions shown in fig-
ure 4.
A third important result is the observation that the
joint activity of grid cell groups that process differ-
ent modalities (Fig. 6) shows a much stronger pattern
separation than the joint activity of grid cell groups
that have different grid spacings (Fig. 5). Yet, the grid
cell literature focuses much more on the latter (Rolls
et al., 2006; Solstad et al., 2006; Franzius et al., 2007;
de Almeida et al., 2009; Savelli and Knierim, 2010),
while aspects of multimodality appear to be underrep-
resented (Aronov et al., 2017; Diehl et al., 2017).
7 CONCLUSION
We investigated the degree to which entorhinal grid
cells perform pattern separation and thus may facil-
itate the subsequent pattern separation observed in
areas of the hippocampus. Based on the hypothe-
sis that the behavior of entorhinal grid cells is just
one instance of a more general information process-
(a) cell #1
(b) cell #10
(c) cell #20
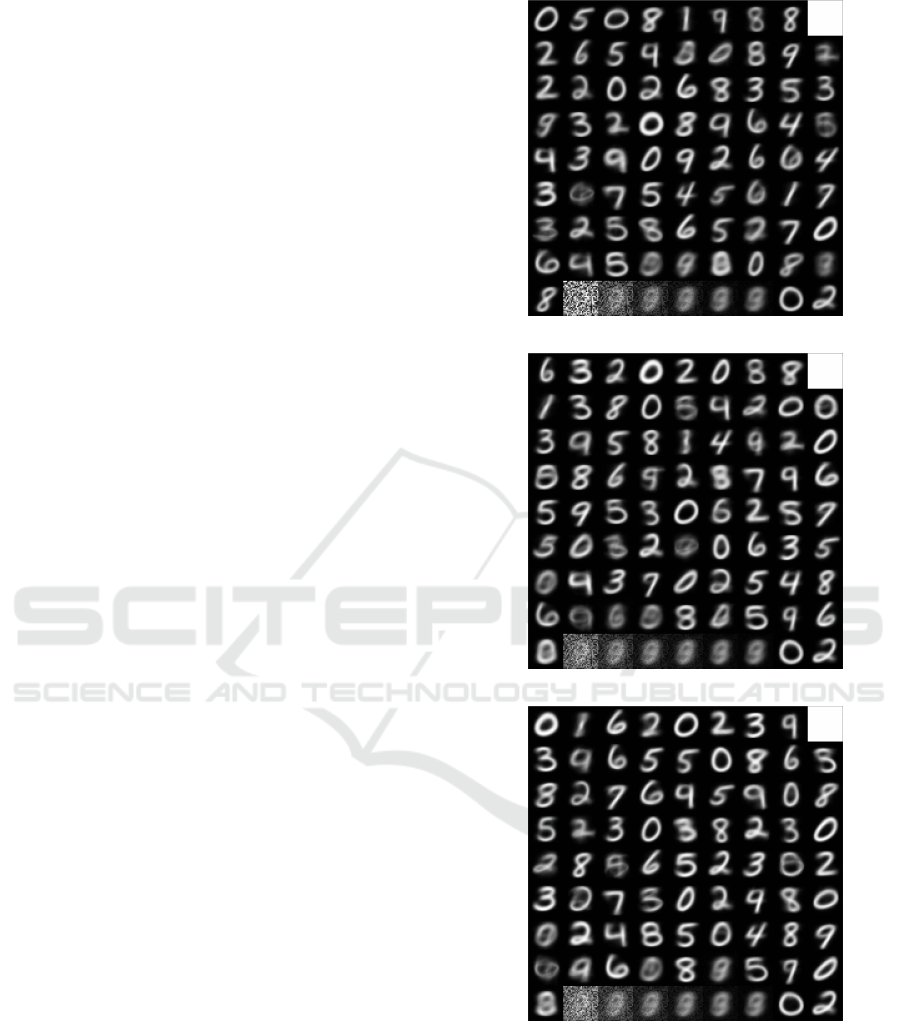
Figure 7: Three examples of 80 prototypes that were
learned by three grid cells {#1,#10,#20} to form an input
space representation. The prototypes typically cover the en-
tire input space in each grid cell and are slightly shifted in
the input space across different cells. The last rows in the
prototype overviews show examples of “unused” prototypes
that the cells haven’t utilized so far.

ing scheme we used our recently introduced RGNG-
based grid cell model (Kerdels and Peters, 2015;
Kerdels, 2016; Kerdels and Peters, 2016) to simulate
several groups of grid cells that processed visual in-
puts derived from the MNIST database of handwritten
digits. The results indicate that entorhinal grid cells
may indeed perform pattern separation on their input
signals and thus may provide already orthogonalized
output signals to the hippocampus.
These results challenge established views on the
parahippocampal-hippocampal region and provide a
novel explanation for the origin of the pattern sepa-
ration that is observed in the dentate gyrus. Further-
more, they uncover a common misconception about
the assumed lack of specificity in the activity of grid
cell groups. Finally, the results encourage the further
investigation of the influence of multimodal input on
the processing within the entorhinal cortex.
REFERENCES
Aronov, D., Nevers, R., and Tank, D. W. (2017). Map-
ping of a non-spatial dimension by the hippocampa-
lentorhinal circuit. Nature, 543(7647):719–722.
Berron, D., Sch
¨
utze, H., Maass, A., Cardenas-Blanco, A.,
Kuijf, H. J., Kumaran, D., and D
¨
uzel, E. (2016).
Strong evidence for pattern separation in human den-
tate gyrus. Journal of Neuroscience, 36(29):7569–
7579.
Burgess, N., Maguire, E. A., and O’Keefe, J. (2002). The
human hippocampus and spatial and episodic mem-
ory. Neuron, 35(4):625 – 641.
Butler, A. and Hodos, W. (2005). Comparative Vertebrate
Neuroanatomy: Evolution and Adaptation. Wiley.
Constantinescu, A. O., O’Reilly, J. X., and Behrens, T.
E. J. (2016). Organizing conceptual knowledge in hu-
mans with a grid-like code. Science (New York, N.Y.),
352(6292):1464–1468.
de Almeida, L., Idiart, M., and Lisman, J. E. (2009). The
inputoutput transformation of the hippocampal gran-
ule cells: From grid cells to place fields. The Journal
of Neuroscience, 29(23):7504–7512.
Diehl, G. W., Hon, O. J., Leutgeb, S., and Leutgeb, J. K.
(2017). Grid and nongrid cells in medial entorhinal
cortex represent spatial location and environmental
features with complementary coding schemes. Neu-
ron, 94(1):83 – 92.e6.
Franzius, M., Vollgraf, R., and Wiskott, L. (2007). From
grids to places. Journal of Computational Neuro-
science, 22(3):297–299.
Fritzke, B. (1995). A growing neural gas network learns
topologies. In Advances in Neural Information Pro-
cessing Systems 7, pages 625–632. MIT Press.
Fyhn, M., Molden, S., Witter, M. P., Moser, E. I., and
Moser, M.-B. (2004). Spatial representation in the en-
torhinal cortex. Science, 305(5688):1258–1264.
Hafting, T., Fyhn, M., Molden, S., Moser, M.-B., and
Moser, E. I. (2005). Microstructure of a spatial map
in the entorhinal cortex. Nature, 436(7052):801–806.
Kerdels, J. (2016). A Computational Model of Grid Cells
based on a Recursive Growing Neural Gas. PhD the-
sis, FernUniversit
¨
at in Hagen, Hagen.
Kerdels, J. and Peters, G. (2015). A new view on grid cells
beyond the cognitive map hypothesis. In 8th Confer-
ence on Artificial General Intelligence (AGI 2015).
Kerdels, J. and Peters, G. (2016). Modelling the grid-like
encoding of visual space in primates. In Proceedings
of the 8th International Joint Conference on Compu-
tational Intelligence, IJCCI 2016, Volume 3: NCTA,
Porto, Portugal, November 9-11, 2016., pages 42–49.
Killian, N. J., Jutras, M. J., and Buffalo, E. A. (2012). A
map of visual space in the primate entorhinal cortex.
Nature, 491(7426):761–764.
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
Leutgeb, J. K., Leutgeb, S., Moser, M.-B., and Moser, E. I.
(2007). Pattern separation in the dentate gyrus and ca3
of the hippocampus. Science, 315(5814):961–966.
Martinetz, T. M. and Schulten, K. (1994). Topology repre-
senting networks. Neural Networks, 7:507–522.
Rolls, E. (2013). The mechanisms for pattern completion
and pattern separation in the hippocampus. Frontiers
in Systems Neuroscience, 7(74).
Rolls, E. T., Stringer, S. M., and Elliot, T. (2006). Entorhi-
nal cortex grid cells can map to hippocampal place
cells by competitive learning. Network: Computation
in Neural Systems, 17(4):447–465. PMID: 17162463.
Rowland, D. C., Roudi, Y., Moser, M.-B., and Moser, E. I.
(2016). Ten years of grid cells. Annual Review of
Neuroscience, 39(1):19–40. PMID: 27023731.
Savelli, F. and Knierim, J. J. (2010). Hebbian analysis of the
transformation of medial entorhinal grid-cell inputs to
hippocampal place fields. Journal of Neurophysiol-
ogy, 103(6):3167–3183.
Solstad, T., Moser, E. I., and Einevoll, G. T. (2006). From
grid cells to place cells: A mathematical model. Hip-
pocampus, 16(12):1026–1031.
Somogyi, P. (2010). Hippocampus: Intrinsic organization.
In Shepherd, G. M. and Grillner, S., editors, Hand-
book of Brain Microcircuits, pages 148–164. Oxford
University Press.
Squire, L., Bloom, F., Spitzer, N., Squire, L., Berg, D.,
du Lac, S., and Ghosh, A. (2008). Fundamental Neu-
roscience. Fundamental Neuroscience Series. Elsevier
Science.
Tulving, E. and Markowitsch, H. J. (1998). Episodic and
declarative memory: Role of the hippocampus. Hip-
pocampus, 8(3):198–204.
van Strien, N. M., Cappaert, N. L. M., and Witter, M. P.
(2009). The anatomy of memory: an interactive
overview of the parahippocampal-hippocampal net-
work. Nat Rev Neurosci, 10(4):272–282.
Witter, M. P., Wouterlood, F. G., Naber, P. A., and van
Haeften, T. (2000). Anatomical organization of the

parahippocampal-hippocampal network. Annals of the
New York Academy of Sciences, 911(1):1–24.
Yassa, M. A. and Stark, C. E. L. (2011). Pattern sepa-
ration in the hippocampus. Trends in neurosciences,
34(10):515–525.
APPENDIX
Parameterization
Each layer of an RGNG requires its own set of pa-
rameters. In case of our two-layered grid cell model
we use the sets of parameters θ
1
and θ
2
, respec-
tively. Parameter set θ
1
controls the main top layer
RGNG while parameter set θ
2
controls all bottom
layer RGNGs. Table 1 summarizes the parameter val-
ues used for the simulation runs presented in this pa-
per. For a detailed characterization of these parame-
ters we refer to Kerdels (Kerdels, 2016).
Table 1: Parameters of the RGNG-based model used
throughout all simulation runs. Parameters θ
1
control the
top layer RGNG while parameters θ
2
control all bottom
layer RGNGs of the model.
θ
1
θ
2
ε
b
= 0.004 ε
b
= 0.001
ε
n
= 0.004 ε
n
= 0.00001
ε
r
= 0.01 ε
r
= 0.01
λ = 1000 λ = 1000
τ = 300 τ = 300
α = 0.5 α = 0.5
β = 0.0005 β = 0.0005
M = 100 M =
{
20, 40, 80
}
