
Medical Image Processing in the Age of Deep Learning
Is There Still Room for Conventional Medical Image Processing Techniques?
Jason Hagerty, R. Joe Stanley and William V. Stoecker
Missouri University of Science and Technology, 1201 N State St., Rolla, MO, U.S.A.
{jrh55c, stanleyj, wvs}@mst.edu
Keywords: Deep Learning, Convolution Neural Networks, Fusion, Transfer Learning.
Abstract: Deep learning, in particular convolutional neural networks, has increasingly been applied to medical
images. Advances in hardware coupled with availability of increasingly large data sets have fueled this rise.
Results have shattered expectations. But it would be premature to cast aside conventional machine learning
and image processing techniques. All that deep learning comes at a cost, the need for very large datasets.
We discuss the role of conventional manually tuned features combined with deep learning. This process of
fusing conventional image processing techniques with deep learning can yield results that are superior to
those obtained by either learning method in isolation. In this article, we review the rise of deep learning in
medical image processing and the recent onset of fusion of learning methods. We discuss supervision
equilibrium point and the factors that favor the role of fusion methods for histopathology and quasi-
histopathology modalities.
1 INTRODUCTION
Because deep learning architectures, in particular the
convolutional neural net (convnet), have attracted
unprecedented attention in medical image
processing, there is a tendency to overlook the
potential contribution of conventional image
processing techniques. The allure of the new
convnet architecture is that it will simplify the task
of image processing. But this convenience comes at
a cost, primarily in demand for more training
examples, and a case will be made here that there is
still a place in image processing for more
conventional computer vision techniques. This
article focuses on the rise of deep learning, in
particular the convnet architecture, the relation
between image complexity and image processing
architecture, and discusses the rule of fusion of
conventional and deep learning architectures. To
better understand the need for conventional learning
techniques, we define two new image complexity
measures. We use these image complexity measures
to define the learning equilibrium (dataset size at
which deep learning techniques gain superiority) as
a function of image complexity. We explore the
situations where fusion of new and conventional
image processing techniques offers the best image
processing solution. Finally, we give examples
where conventional learning and deep learning
fusion has already proven successful.
2 THE RISE OF DEEP LEARNING
IN IMAGE PROCESSING
Deep learning (representation learning) computa-
tional models comprise a sequence of processing
layers operating independently on numeric data to
independently learn hierarchical data representations
(LeCun, 2015; Bengio, 2013; Goodfellow, 2016).
Deep learning can discover intricate structures in
large data sets by using the backpropagation
algorithm to indicate how a machine should change
its internal parameters. Since deep learning
architecture encompasses layers of nodes updating
operating parameters in sequence, it is a type of
neural network. Deep learning models differ from
other neural networks by using a deep graph with
multiple processing layers of a small number of
nodes, as opposed to traditional neural networks,
comprised of few layers with a larger number of
nodes (LeCun, 2015; Bengio, 2013; Goodfellow,
2016). Deep learning, as the term “representation
learning” implies, seeks to discover knowledge
representations rather than to use hand-crafted
knowledge representations. In the past decade, the
306
Hagerty J., Stanley R. and Stoecker W.
Medical Image Processing in the Age of Deep Learning - Is There Still Room for Conventional Medical Image Processing Techniques?.
DOI: 10.5220/0006273803060311
In Proceedings of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2017), pages 306-311
ISBN: 978-989-758-225-7
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

use of the phrase “deep learning” has exploded. A
search on IEEE Xplore returns only 36 articles
published in 2006 vs. 1,017 articles for 2015. The
increasing use of deep learning in research can be
attributed to advances in several areas: the
development of large data sets, also called “big
data,” a dramatic increase in computational power,
and the desire to “re-brand” neural networks,
echoing earlier efforts to rebrand “artificial
intelligence” and “artificial neural networks” (Allen,
2017; LeCun 1998).
Figure 1: Left to right: a.) Input layer accepting a 32x32
RGB image. b.) convolution consisting of 8 7x7 filters. c.)
2x2 max-pooling layer. d.) Fully connected layer with
1536 input nodes for each pixel from the previous layer
and 256 output nodes. e.) Fully connected layer consisting
of 256 inputs nodes plus a bias node and a single sigmoid
activated output node. Total number of free parameters:
525,985.
One deep learning architecture that has been
prominently successful in image recognition
challenges (Goodfellow, 2015) is the convolutional
neural network (convnet). The basic convnet
architecture combines two concepts: the
mathematical convolution operator and a fully
connected neural network. One or more convolution
layers are usually prepended to a fully connected
network. A simple convnet with a single convolution
layer is presented in Figure 1. The application of the
2D convolution operator, shown in Equation 1,
within the convolution layer, enables the network to
process an input image directly without the need of
“flattening” the image, preserving any spatial
relations that may exist in the image. The convnet
architecture was introduced in 1998 when LeCun
presented LeNet (LeCun, 1998) designed to identify
handwritten digits; LeNet yielded a remarkably low
error rate of 0.7%. Equation (1) summarizes the
operation of a kernel k(x,y) upon an image I(x,y).
,
∗
,
,
,
(1)
The addition of convolution layers to a typical
neural network and allowing the back-propagation
training algorithm to update not only the weights of
the fully connected neural network but also the
elements of the 2D convolution kernels, allows the
convnet to directly use images as inputs and
alleviates the need to manually determine the “best”
convolution kernel. Throughout the training period,
the “best” convolutional kernel will be continually
improved upon. This has enabled investigators to
focus on optimizing the architecture of the network
(machine learning) without requiring conventional
manually-tuned feature extraction (computer vision).
But this convenience comes at a cost, the number
of weights (parameters) in the convnet is large and
presents a computational burden on the
backpropagation training algorithm. For the simple
network shown in Figure 1, a total of 525,985
parameters need tuning! The high computational
demands to optimize that many parameters is not the
only concern; because of the parameter count, a
large number of training samples is required for
successful training and generalization.
The high computational requirement to be able to
use deep learning has been somewhat alleviated by a
dramatic increase in computational power from the
now common use of multiple cores (CPUs) in
current processors and specialized graphics
processing units (GPUs). The GPU came about
because of the demands of computer game players
for more detailed graphics. Rendering a scene for a
computer game requires many floating-point matrix
operations. The developers of these GPUs designed
these processors to include hundreds, sometimes
thousands, of cores that are specifically designed to
perform fast and efficient floating-point matrix
operations in an effort to offload the burden from the
CPU. An unexpected but welcomed result was that
the GPUs could be harnessed for machine learning,
since neural networks could also be expressed in a
sequence of floating point matrix operations.
Machine learning algorithms began to be
developed and implemented in a parallel manner to
take advantage of these GPUs. These parallel
algorithms could now be leveraged on clusters of
CPUs or ideally, GPUs. As a result of parallel
implementation of machine learning algorithms
along with fast floating-point operation via use of
GPUs, a 9x reduction in training times is possible
when comparing a single GPU to the single CPU
processor with multiple cores (Brown, 2015).
3 DATA IS THE PROBLEM
The computational demands of deep learning
Medical Image Processing in the Age of Deep Learning - Is There Still Room for Conventional Medical Image Processing Techniques?
307
algorithms are mostly addressed with use of GPUs,
but the number of parameters that require
optimization in a deep learning algorithm is still a
problem. Because of the number of parameters,
training and generalization demand a large training
set. In several domains, publically available large
datasets exist, for example, the ImageNet dataset.
The ImageNet dataset has over 14 million images
that encompass 14 thousand classes (ImageNet,
2016). But for domains such as medicine, although
datasets of moderate size are increasingly available,
very large datasets on the order of ImageNet are not
available. The largest dermoscopy image set, for
example, is located at the ISIC project (ISIC, 2016)
and consists of approximately 12 thousand images,
but only about 700 of those are of melanoma.
Because of the relatively small number of images,
and the heavily biased number of one class (benign
versus melanoma), researchers cannot blindly use a
deep learning algorithm and expect good results.
To use a deep learning approach with the ISIC
dataset, one should augment the original dataset by
including rotated, flipped and mirrored versions of
the original images. Oversampling the minority class
can be used to minimize the bias between classes. A
researcher may also use a network trained in another
domain, such as ImageNet or AlexNet, and use a
technique called transfer learning to train a new
network using the combined features of the pre-
trained network and new features specific to the
learning task. Or a researcher may have to rely on
manually tuned feature extractors to create an input
vector to a learning algorithm that is not a complex
deep learning algorithm.
With smaller datasets, a convnet may not have
the optimal solution architecture. For some domains,
large image sets may not be available. For example,
for skin lesions, the image datasets available may
only contain 10’s or 100’s of examples of a
particular lesion diagnosis. In the future, larger
image sets may become available, as anticipated for
the ISIC project. But these datasets still require
professionals to collect, label and curate the data
accurately and still may only increase by an order or
two of magnitude.
This is where conventional image processing
may continue to excel. Since the image datasets in
specialized fields are usually quite small, manual
extraction of dominant features will offset the lack
of data. These critical features are often the same
features that professional look for in the clinic.
4 MORE COMPLEX IMAGE
SETS REQUIRE MORE
IMAGES FOR SUCCESSFUL
CLASSIFICATION
Recent image recognition challenges, such as those
using the ImageNet dataset (Figure 2), may include
images with varying scenes at different scales and
containing multiple objects. An index of image
complexity (i.c.) can be defined for an image.
Additionally, image complexity can be defined for
an entire set of images. An image complexity index
should be higher if 1) image object sizes vary widely
in scale 2) multiple objects are present in the image
3) distracting objects are present in the image. An
image set is more complex if 1) average image
complexity is higher 2) more classes of images are
present and 3) inter-image variety within a class is
greater. Thus the ImageNet dataset, with various
complex scenes, is quite complex and is quite large.
Intuitively, we may suppose that larger image
datasets are needed for successful diagnosis of more
complex image sets.
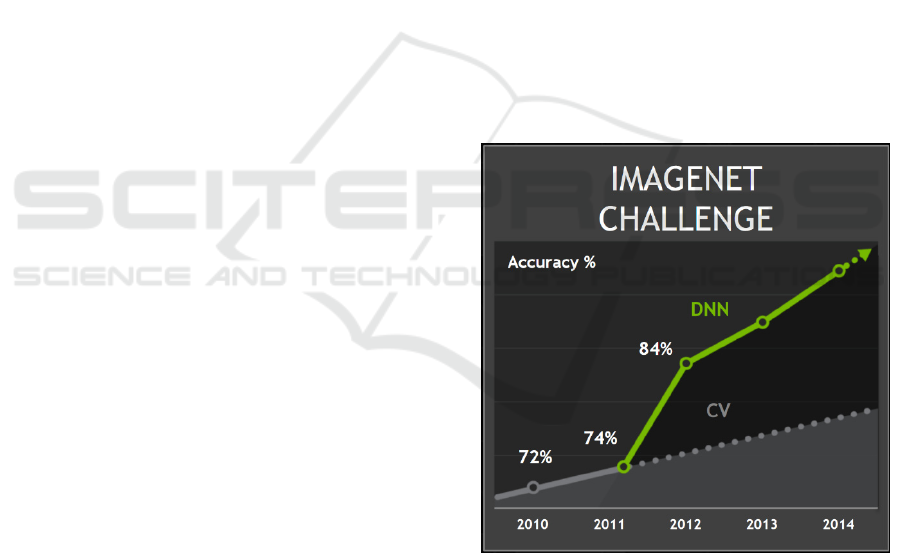
Figure 2. ImageNet challenge result. Beginning in 2011,
deep learning (DNN) results (solid line), began to surpass
those obtained from traditional learning (dashed line).
(Brown, 2015).
5 IMAGE COMPLEXITY AND
THE SUPERVISION
EQUILIBRIUM
Previous sections establish that deep learning techni-
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
308
ques need large datasets before accuracy exceeds
that of conventional techniques. The size of the
dataset needed for successful classification is
expected to grow as images and image sets increase
in complexity. Let us consider the case number
spectrum, ranging from low numbers of cases to
very high numbers of cases. We plot the number of
cases on a log scale, shown in Figure 3. In some
two-class problems, such as benign vs. malignant
diagnosis, small datasets may contain equal numbers
of images of benign and malignant cases. For the
zero-knowledge situation, over many trials, as with
coin flipping, we expect 50% diagnostic accuracy.
As the number of cases grows, the expected
accuracy of both conventional and deep learning-
based models tends to increase, with conventional
learning accuracy higher for a small number of
cases. As case numbers grow, at some point, deep
learning techniques become equal in classification
accuracy to conventional techniques, as shown in
Figure 3.
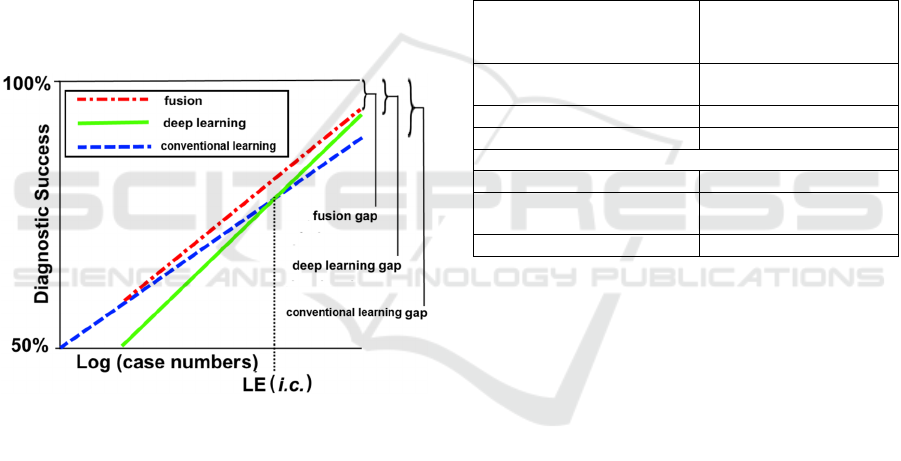
Figure 3: Our conjectured model of diagnostic success for
conventional learning techniques (dashed line), deep
learning techniques (solid line), and fusion techniques
(dot-dashed line). Errors (learning gaps) persist, even with
large case numbers, for all three techniques. Curve
shapes, shown here as linear functions of log (case
numbers), are unknown.
We may define this equilibrium point, where
both deep and conventional learning have the same
diagnostic accuracy as the Learning Equilibrium
(LE). LE is a function of image complexity (i.c.).
Different image spaces have different levels of
complexity, due to both intra- and inter-image
complexity, as noted above. As image space
complexity grows, the number of images required to
represent that complexity grows; the accuracy
obtained for any given number of cases falls. Thus
for high complexity image sets, the accuracy curves
flatten, and; LE grows. We offer the conjecture that
LE(i.c.), with appropriate smoothing, is a
monotonically increasing function of i.c..
As shown in Figure 3, diagnostic success can
never be perfect. Errors persist, even with large case
numbers, due to imperfect knowledge of the image
space. These gaps in knowledge in the three
representations—the conventional learning gap, the
deep learning gap, and the fusion gap, all become
relatively smaller as the number of cases increases,
but will always remain nonzero. In the real world,
perfect diagnostic accuracy remains elusive. Even
histopathologic “gold standards” have an inherent
degree of uncertainty. Expert pathologists disagree
on diagnoses (Krieger, 1994). This creates a
challenge in image machine learning (Guo, 2015).
Table 1: Conventional vs. Deep Learning.
Elements favoring
conventional machine
learning
Examples
Repeating biological units
Cells, nuclei in
histopathologic images
Scale invariance of features Vessel walls
High domain knowledge Organs e.g. brain, heart
Elements favoring DL Examples
No repeating units
Microcalcifications in
breast cancer
Features vary in scale Bone tumors
6 LOW HANGING FRUIT FOR
FUSION TECHNIQUES
Table 1 shows types of elements in medical images
favoring either conventional learning or deep
learning. Types of images favoring conventional
learning include images with repeating biological
units as seen in histopathology, scale-invariant
images as seen in vessel walls, and organs such as
brain and heart described with specific domain
knowledge. In these areas, human-supervised
conventional learning can add significant
information to deep learning by adding biological
descriptions which successfully constrain class
output. Thus we predict that human-supervised
conventional learning will continue to be useful in
histopathology, brain and cardiovascular imaging.
We may also predict that quasi-pathological
domains using newer techniques such as
dermoscopy, confocal microscopy and optical
coherence tomography (OCT) may also utilize
conventional techniques, in some cases fused with
Medical Image Processing in the Age of Deep Learning - Is There Still Room for Conventional Medical Image Processing Techniques?
309

deep-learning techniques for some time to come.
Deep learning, in contrast, is already showing
progress in automated unsupervised analysis of
mammograms (Suzuki, 2016; Wang, 2016),
Three deep-conventional learning fusion
examples have already appeared in the field of
automated histopathology. Zhong and colleagues
fused information from deep learning and
conventional learning (Zhong, 2017). In comparing
multiple machine learning strategies, it was found
that the combination of biologically inspired
conventional cellular morphology features (CMF)
and predictive sparse decomposition deep learning
features provided the best separation of benign and
malignant histology sections (Zhong, 2017). The
deep learning arm used a pre-trained AlexNet
network (transfer learning). The conventional arm
used cellular morphology features, which include
nuclear size, aspect ratio, and mean nuclear gradient.
The researchers concluded that both CMF features
and sparse decomposition deep learning features
encode meaningful biological patterns.
Wang and colleagues were able to detect mitoses
in breast cancer histopathology images by using the
combined manually-tuned CMF data and convolu-
tional neural net features (Wang, 2014).
Arevalo and colleagues added an interpretable
layer they called “digital staining,” to aid in their
deep learning approach to classification of basal cell
carcinoma (Arevalo, 2015). Of interest, the
handcrafted layer finds the area of importance,
reproducing the high-level search strategy of the
expert pathologist.
7 CONCLUSION
Deep learning has shown its ability to solve, with a
high degree of accuracy, rather complex problems.
But conventional machine learning and image
processing techniques should not be totally
discounted. Deep learning’s ability does not come
without a cost: time and dataset requirements. With
very large datasets, deep learning is already the
preferred method to use, but may not be ideal for
smaller datasets. Although conventional machine
learning and image processing may be more labor
intensive, they provide a tool for situations lacking
sufficient data, despite augmentation techniques. We
offer a conjectural model which shows advantages
for conventional learning techniques for small
datasets; advantages shift to deep learning after
some dataset size. We call this dataset size the
“learning equilibrium” (LE). It would be interesting
to study how many images are needed for deep
learning approaches to be effective in different
applications. Another topic for future research is to
determine the characteristics that make one
application require a larger dataset than another. We
may consider the dataset size at the LE to be an
application-specific trade-off; for applications in
which conventional models are effective, the LE
point will be larger.
In some applications, such as histopathology, and
related applications such as dermoscopy, biological
constraints are best modeled by manually-tuned
features. Therefore in these applications especially,
the LE dataset size is large. In these applications
there is still room for familiar computer vision
techniques in the novel world of deep learning.
REFERENCES
LeCun Y., Bengio Y., Hinton G. Deep learning. Nature.
2015 May 28;521(7553):436-44.
Bengio Y., Courville A., Vincent P. Representation
learning: a review and new perspectives. IEEE Trans
Pattern Anal Mach Intell. 2013 Aug; 35(8):1798-828.
Goodfellow I., Bengio Y., Courville A. Deep Learning.
Cambridge MA, MIT Press, 2016.
Allen, Kate. "How a Toronto Professor's Research
Revolutionized Artificial Intelligence | Toronto Star."
Thestar.com. N.p., 17 Apr. 2015. Web. 09 Jan. 2017.
LeCun Y., Bottou L., Bengio Y., Haffner, P., Gradient-
Based Learning Applied to Document Recognition,
Proceedings of the IEEE, 86(11):2278-2324, Nov.
1998.
Goodfellow I. J., Erhan D., Luc Carrier P., et al.
Challenges in representation learning: a report on three
machine learning contests. Neural Netw. 2015 Apr;
64:59-63. PMID: 25613956
Valiant L. "A theory of the learnable", Commun. ACM,
vol. 27, pp. 1134-1142, Nov. 1984.
Brown L. “Deep learning with GPUs”, Larry Brown
Ph.D., Johns Hopkins University, June 2015
http://www.nvidia.com/content/events/geoInt2015/LB
rown_DL.pdf accessed November 28, 2016
"ImageNet", Image-net.org, 2016. [Online]. Available:
http://image-net.org/. [Accessed: 29- Nov- 2016].
"ISIC Archive", Isic-archive.com, 2016. [Online].
Available: https://isic-archive.com. [Accessed: 29-
Nov- 2016].
Krieger N., Hiatt R. A., Sagebiel R. W., Clark W. H.,
Mihm M.C. Inter-observer variability among
pathologists' evaluation of malignant melanoma:
effects upon an analytic study. J Clin Epidemiol. 1994
Aug; 47(8):897-902.
Guo P., Banerjee K., Stanley R., Long R., Antani S.,
Thoma G., Zuna R., Frazier S., Moss R., Stoecker W.
Nuclei-Based Features for Uterine Cervical Cancer
Histology Image Analysis with Fusion-based
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
310

Classification. IEEE J Biomed Health Inform. 2015
Oct 26. [Epub ahead of print]
Suzuki S., Zhang X., Homma N., Ichiji K., Kawasumi Y.,
Ishibashi T., Yoshizawa M. WE-DE-207B-02:
Detection of Masses On Mammograms Using Deep
Convolutional Neural Network: A Feasibility Study.
Med Phys. 2016 Jun; 43(6):3817.
Wang J., Yang X., Cai H., Tan W., Jin C., Li L.
Discrimination of Breast Cancer with Microcalcifica-
tions on Mammography by Deep Learning. Sci Rep.
2016 Jun 7; 6:27327.
Zhong C., Han J., Borowsky A., Parvin B., Wang Y.,
Chang H. When machine vision meets histology: A
comparative evaluation of model architecture for
classification of histology sections. Med Image Anal.
2017 Jan; 35:530-543.
Wang H., Cruz-Roa A., Basavanhally A., Gilmore A. H.,
Shih N., Feldman M., et al. Mitosis detection in breast
cancer pathology images by combining handcrafted
and convolutional neural network features. J Med
Imaging, 1 (3) (2014), p. 034003
Arevalo J., Cruz-Roa A., Arias V., Romero E., González
F.A. An unsupervised feature learning framework for
basal cell carcinoma image analysis. Artif Intell Med.
2015 Jun; 64(2):131-45.
Medical Image Processing in the Age of Deep Learning - Is There Still Room for Conventional Medical Image Processing Techniques?
311
