
An Image Generator Platform to Improve Cell Tracking Algorithms
Simulation of Objects of Various Morphologies, Kinetics and Clustering
Pedro Canelas
1
, Leonardo Martins
1
, André Mora
1
, Andre S. Ribeiro
2
and José Fonseca
1
1
Computational Intelligence Group of CTS/UNINOVA, Faculdade de Ciências e Tecnologia,
Universidade Nova de Lisboa, Quinta da Torre, 2829-516, Caparica, Portugal
2
Laboratory of Biosystem Dynamics, Dep. of Signal Processing, Tampere University of Technology, Tampere, Finland
Keywords: Microscopy, Synthetic Time-lapse Image Simulation, Cell Tracking, Cluster Tracking.
Abstract: Several major advances in Cell and Molecular Biology have been made possible by recent advances in live-
cell microscopy imaging. To support these efforts, automated image analysis methods such as cell
segmentation and tracking during a time-series analysis are needed. To this aim, one important step is the
validation of such image processing methods. Ideally, the “ground truth” should be known, which is
possible only by manually labelling images or in artificially produced images. To simulate artificial images,
we have developed a platform for simulating biologically inspired objects, which generates bodies with
various morphologies and kinetics and, that can aggregate to form clusters. Using this platform, we tested
and compared four tracking algorithms: Simple Nearest-Neighbour (NN), NN with Morphology and two
DBSCAN-based methods. We show that Simple NN works well for small object velocities, while the others
perform better on higher velocities and when clustering occurs. Our new platform for generating new
benchmark images to test image analysis algorithms is openly available at
(http://griduni.uninova.pt/Clustergen/ClusterGen_v1.0.zip).
1 INTRODUCTION
Recent advances in live-cell microscopy imaging
have enabled the acquisition of images with higher
quality and resolution and the development of
techniques for detecting and observing recently
discovered cellular structures and their kinetics
(Danuser, 2011; Sung and McNally, 2011).
The main challenges in live-cell imaging can be
divided into two areas. The first relates to processes
that occur before and during image acquisition in the
microscope, associated with the refinement of
processes, such as illumination, focus, drift
correction, stage positioning and refinement of
microscope components (e.g. shutter, lens, camera,
stage) (Coutu and Schroeder, 2013). The second is
related to post processing limitations, associated
with storage of large amounts of data and image
processing (e.g., image registration, segmentation,
tracking, statistical quantification and background
correction) (Coutu and Schroeder, 2013; Bonnet,
2004).
Automatic correction algorithms are normally
included in microscope software packages (Frigault
et al., 2009), while the process of image registration
(overlaying two or more images of the same location
taken at different time frames and/or from different
viewpoints and/or by different sensorial devices) has
been extensively studied and several methods are
available. These are classified based on modality,
intensity, type of data, dimensionality, domain and
type of transformation, and registration
methodologies (Deshmukh and Bhosle, 2011;
Wyawahare et al., 2009).
The next step is related to the segmentation of
cells or cellular structures of interest (Meijering
2012), where these segmented objects are detected,
located, and separated from the background. The
main challenge is to automatize this process and
provide it with a high specificity and sensitivity for a
vast number of cases. Presently, most algorithms are
made available in open-source platforms and apply
several approaches, such as intensity thresholding,
feature detection, morphological filtering, region
accumulation, deformable model fitting, etc.
(Meijering, 2012).
When handling a time series, one needs to link
the segmented objects in the actual frame with the
ones from the previous frame, so as to extract the
44
Canelas, P., Martins, L., Mora, A., Ribeiro, A. and Fonseca, J.
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and Clustering.
DOI: 10.5220/0005957800440055
In Proceedings of the 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH 2016), pages 44-55
ISBN: 978-989-758-199-1
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

object’s trajectory over time. Having available the
information describing the target, defined by the
state sequence X
k
, kϵ
(where
is the set of
frames), and the measurements defined by Z
k
, the
objective of tracking is to estimate X
k
, given all
measurements until the moment Z
1:k
(Tissainayagam
and Suter, 2005). This is made difficult by noise,
occlusions, illumination changes, complex motions
and object’s shape dynamics, which can enhance the
misidentification of object tracks over time (Yilmaz
et al., 2006).
Presently available tools for segmentation and
tracking in different microscopy settings include the
‘Cell-C’, based on DAPI staining and fluorescence
in situ hybridization images (Selinummi et al.,
2005), ‘CellTracer’, which applies morphological
methods to automatically segment bacterial cells,
yeast and human cells (Wang et al., 2010),
‘MicrobeTracker’ and its accessory tool
‘SpotFinder’, which segment Escherichia coli and
Caulobacter crescentus cells and detected
fluorescent spots within (Sliusarenko and Heinritz,
2011), ‘Schnitzcells’, which segments and tracks E.
coli cells in confocal or phase contrast images
(Young et al., 2012) and, ‘CellAging’, which was
developed for cell segmentation and tracking in
order to study the segregation and partitioning in cell
division of protein aggregates (Häkkinen et al.,
2013).
Validation of these image processing tools and
techniques they use require the use of gold-standard
images, usually manually annotated by biology
experts. This is problematic, as it is expert-
dependent (both inter-user and intra-user variability
can be high) and are impractical in high-throughput
data-sets (Coelho et al., 2009). To overcome this
problem, a viable alternative is to generate artificial
images using biologically inspired models. These
images, whose ground truth is known, can be used
for the accurate quantitative evaluation of image
processing algorithms (Bonnet, 2004).
In Section 2, we give a comprehensive literature
review of existing tools for simulation of synthetic
microscopy images and the recent developments on
cell tracking algorithms. In Section 3 we present the
contributions to the development of the image
simulation tool (models and parameters) and the
implementation of three different tracking
algorithms. In Section 4 the tracking results of
several examples are presented using different
parameters. Finally, in Section 5 we present final
remarks on the development of the simulation tool
and the results of the three algorithms, along
potential future endeavours.
2 STATE OF THE ART
2.1 Synthetic Image Generators
There have been several contests and open
challenges, usually requiring that each methodology
is tested on the same benchmark data-sets (acquired
by an independent laboratory or created by artificial
image generators) (Meijering, 2012). Such artificial
image generators require realistic biological models,
and commonly use theoretical and experimental
information regarding the statistical distributions of
the object’s behaviour (Xiong et al., 2010) but also
spatial and temporal data from the object (Kruse,
2012; Misteli, 2007). If the object studied is a cell,
these models should include morphology parameters
such as cell shape and size, location of subcellular
structures, kinetic and spatial statistics of cell
growth, cell division, cell migration and models of
internal cell functions.
The modus operandi of presently available tools
to simulate microscopy images based on biological
models can be divided in three stages: the digital
phantom object generation, the simulation of the
signal passing through the optical system and, the
simulation of the image formed on a specific sensor
(Svoboda et al., 2009).
Simulators such as ‘SIMCEP’ (Lehmussola et
al., 2007) have provided a gold-standard platform to
validate and test various image processing tools,
such as the previously mentioned ‘CellC’
(Selinummi et al., 2005), the open-source and Java-
based image processor ImageJ, and the
commercially available MCID Analysis (Imaging
Research Inc., Catharines, ON, Canada; Evaluation
ver. 7.0), along with other image processing tools
(Ruusuvuori et al., 2008). The phantom objects are
generated with different cell parameters, such as
probability of clustering, cell radius, and cell shape
and with parameters related to the sensors and the
optical system such as background noise and
illumination disturbance (Ruusuvuori et al., 2008;
Lehmussola et al., 2011).
Another toolbox, ‘CytoPacq’, was developed
specifically to simulate all three phases, by being
equipped with three different modules. The first
module (‘3D-cytogen’) generates the digital object
phantom, which imitates the cell structure and
behaviour and can generate microspheres,
granulocytes, HL-60 Nucleus and images of Colon
Tissue. The second module (‘3D-optigen’) simulates
the transmission of the signal through the lenses,
objective, excitation filter and emission filter
(various sets of equipment can be simulated). The
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
45

last module, ‘3D-acquigen’ is the digital CCD
camera simulator of the phenomenons that occur
during image capture (noise, sampling, digitization)
by changing the camera selection, the acquisition
time, the dynamic range usage and the stage z-step
(Svoboda et al., 2007; Svoboda et al., 2009). The
same group also introduced a novel versatile tool
(‘TRAgen’), capable of generating 2D time-lapses
by simulating live cell populations as a ground-truth
for the evaluation of cell tracking algorithms. They
include models of cell motility, division and
clustering up to tissue-level density (Ulman et al.,
2015). Both simulators have been an important step
in the simulation of cellular dynamics, such as
measuring protein or RNA levels or even observing
cell migration, division and growth (Coutu and
Schroeder, 2013; Sung and McNally, 2011).
A recently developed toolbox called ‘SimuCell’
(Satwik et al., 2012) is capable of generating
artificial microscopy images with heterogeneous
cellular populations and diverse cell phenotypes.
Each cell and their organelles are modelled with
different shapes, having distinct distributions of
biomarkers over each shape, which can be affected
by the cell’s microenvironment, showing the
importance of good cell placement (e.g. in clusters,
overlapping existing cells) (Satwik et al., 2012).
The ‘CellOrganizer’ toolbox was developed
using a different approach, collecting laboratory data
and using machine-learning techniques to generate
the entire cell, including structures such as the
nucleus, proteins, cell membrane and cytoplasm
components (Murphy, 2012). Although the learn-
based model was capable of extracting a very
precise shape model, it could not be described it in
precise mathematical terms (Zhao and Murphy,
2007).
Most image generators have focused on the
simulation of morphological features and spatial
information of the cell. Morphological information
can suffice to create multidimensional images, but it
cannot simulate time-lapsed multimodal and
functional images, where important time-dependent
processes are present. To simulate such images of
bacterial cells, the ‘miSimBa’ (Microscopy Image
Simulator of Bacterial Cells) tool has been under
development (Martins et al., 2015). The simulated
images can reproduce spatial and temporal bacterial
time-dependent processes by modelling cell growth,
division, motility and morphology: shape, size and
spatial arrangement (Martins et al., 2015).
Relevantly, these simulation tools can also be used
to generate “null-models” (Gotelli and McGill,
1996) to study statistical patterns in absence of a
particular mechanism (e.g. removing the nucleoid to
study how it influences the spatial distribution of
protein aggregates).
2.2 Cell Tracking
Several tracking methods have been proposed,
differing mainly in how to process the available
object features and on the type and number of
tracked objects (Yilmaz et al., 2006). In order to
decide which approach to follow, the object’s
representation, defined during the segmentation
process, must be taken into account. Objects can be
represented through points, geometric shapes,
silhouette and contour, articulated shape model or
skeletal model, leading to the development of
different approaches (Yilmaz et al., 2006). In the
same review, tracking methodologies were divided
into three main categories: Point Tracking, Kernel
Tracking and Silhouette Tracking. Objects in Point
Tracking are represented by points and tracked
based on their position and motion. The main issues
of this methodology are the presence of occlusions
and the entries and exits of objects from the field of
view. This category has been split in Deterministic
and Statistical methods. Deterministic methods
associate each object with the application of motion
constraints, while statistical methods take into
account random perturbations and noise during the
tracking process (Yilmaz et al., 2006). Nearest
neighbour (NN) is the source of all deterministic
approaches and uses only the distances between
objects in k and k-1 matching the objects with the
smallest distances. This distance can be based on
position, shape, colour and size (Elfring et al., 2010).
An efficient visual object tracking algorithm was
proposed by (Gu et al., 2011) that combines NN
classification with descriptors based on the scale-
invariant feature transform, efficient sub-window
search and an updating and pruning method to
achieve balance between stability and plasticity.
This method successfully handles occlusions, clutter,
and changes in scale and appearance.
The probabilistic data association filter (PDAF)
and the joint probabilistic data association filter
(JPDAF) are the basis for the statistical methods.
PDAF uses a weighted average of the measurements
as input, modelling only one target and considering
linear dynamics and measurement models. JPDAF is
an extension of PDAF, allowing multiple target
tracking. The assumptions are the same, calculating
the target’s association probabilities jointly. In both
methods, if the model is linear then the Kalman
Filter has a relevant influence. One of the problems
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
46

of these methods is the incapacity to recover from
errors, because only the last measurement is used
(Elfring et al., 2010). The Kalman filter is an
optimal estimator, which means that it assumes
parameters from indirect, inaccurate and uncertain
observations and if all noise is Gaussian, the linear
Kalman filter minimizes the mean square error of
the estimated parameter. This filter is widely used to
obtain the optimal state estimate (Elfring et al.,
2010).
A different method (Gorji and Menhaj, 2007)
combining the JDPAF and a particle filtering
(Smith, 1993) was proposed and was named ‘Monte
Carlo JPDAF’. This method uses three models: the
first with near constant velocity, the second with
near constant acceleration and a third with both
models, which achieved the best performance.
Another statistical method is the multiple
hypothesis tracking (MHT), which is one of the most
used with point features, but has computational
limitations both in time and memory (Tissainayagam
and Suter, 2005). This method postpones data
association until enough information is available.
The MHT starts by formulating all possible
hypotheses, which develop into a set of new
hypotheses each time new data arrives, generating a
tree of hypothesis (Elfring et al., 2010). For each
hypothesis, the position of the object in the next
frame is predicted and then compared with the
measurements, calculating their distance. The
associations are made for each hypothesis,
generating new hypotheses for the next iteration
(Yilmaz et al., 2006). The tree of hypotheses should
be cut, because it grows exponentially with the
measured data. This can be done by clustering, i.e.,
measurements are subdivided into independent
clusters. If a measurement cannot be associated with
an existent cluster, a new one is created. Another
way of cutting the tree is pruning, meaning that as
new iterations are added, a part of the tree is deleted
(Elfring et al., 2010).
Unlike PDAF and JPDAF, the MHT method can
deal with objects entering, exiting and being
occluded from the field of view. Kernel Tracking
can be done using templates and density-based
appearance models or multi-view appearance
models. Templates use basic geometric shapes,
while multi-view models encode different views of
the object (Yilmaz et al., 2006). Mean shift and KLT
(Kenade-Lucas-Tomasi) are examples of template
and density-based appearance models.
In mean shift, the appearance of objects being
tracked is defined by histograms. Similarities are
measured using the Bhattacharyya coefficient
(Bhattacharyya, 1943) and the Kullback-Leibler
divergence (Joyce, 2014). The process tries to
increase similarity between histograms, by repeating
each iteration until they converge (Zhou et al.,
2009).
KLT is an optical-flow method, which uses
vectors to show the changes in the image (i.e.
translation). A version of this method was proposed
in which the translation of a region centred on an
interest point is iteratively computed. Then, the
tracker evaluates the tracked patch, computing a
transformation between the corresponding patches in
consecutive frames (Shi and Tomasi, 1994). These
methods are effective while tracking single objects,
but have problems dealing with multiple objects.
Silhouette Tracking consists in using precise
information about the shape of the objects, using
Shape Matching and searching for an object
silhouette and its model in each frame. Each
translation from frame to frame is handled separately
by finding corresponding silhouettes detected in two
consecutive frames. Another approach is based on
the evolution of the object contour, connecting the
correspondent objects by state space models or by
minimizing the contour energy (Yilmaz et al., 2006).
When tracking objects, one usually obtains multiple
measurements and the incorrect ones are referred to
as false measurements or clutter. The measurement
with highest probability of being originated from the
tracked object is then selected. If the algorithm
selects the wrong measurement or if the correct
measurement is not detected, a poor state is
estimated. To solve this issue (reducing the
computational cost), a validation region
(measurement gate) is selected. The measurement
gate is a region in which the next measurement has a
higher emergence probability (Elfring et al., 2010).
3 METHODOLOGIES
3.1 Implementation of the Image
Generator - Tool Interface and
Basic Functionalities
The image generator interface and the tracking
methods were implemented using the C# language
from Visual Studio 2015. This sub-section focuses
on the implementation of the image generator and
the basic features. An intuitive and easy to
understand interface was designed in order to
facilitate the analysis of the tracking algorithms. The
time-series generator allows the user to change a set
of settings such as the number of objects, frames,
clusters, and their features. The generator creates a
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
47

csv file for each of the object’s properties (position
in x and y coordinates and a shape-related factor
called “morphology”, which is a rational number
between 0 and 1). In this generator, objects are
represented by circles and the morphology factor is
assumed to be the radius. There is a conversion
factor that determines the maximum radius of the
objects (corresponding to morphology value 1). By
default, this factor is initialized at 30. More factors
and parameters can be added to the algorithm,
increasing its complexity.
The tool interface is shown in Figure 1. At the
top row of the window there are frame handlers, to
advance forward and backward in the time-series, or
to go directly to a specific frame. The “Time-Lapse”
button reproduces the full time-series, one frame per
40 milliseconds.
The left bar contains the boxes to write the
desired width and height of images, in pixels. Then
the user can choose the number of objects in each
frame, and the total number of frames. The
“Maximum Velocity” is the maximum distance, in
pixels, that an object can travel between frames,
while the “Maximum Morphology Difference” is the
maximum difference of the “morphology” factor
that an object can have between frames, in
percentage. The “Physical Move” button controls the
option of giving objects physical limitations to their
kinetics. If it is selected, each object has a velocity
and orientation assigned to it, meaning that its
position dynamics will depend on these two
variables. If it is not selected, objects will move
arbitrarily between frames.
One can also select “Allow Entries/Exits”, to
allow the objects to enter and exit the image limits.
If unselected, objects collide with the edges of the
image when reaching them.
Overlapping of cells is possible, using the
“Allow Occlusions” option. When this option is
selected, objects move without restrictions due to
superposition between them. If it is not selected,
objects will collide between them similarly as when
colliding with the edges.
One can also force the objects to organize into
clusters, by checking the “Create Clusters” option.
When selected, all objects of each cluster have the
same physical features. In this setting, “Physical
Move” is automatically selected and “Allow
Occlusions” is deselected, blocking the
correspondent checkboxes. The button “Cluster
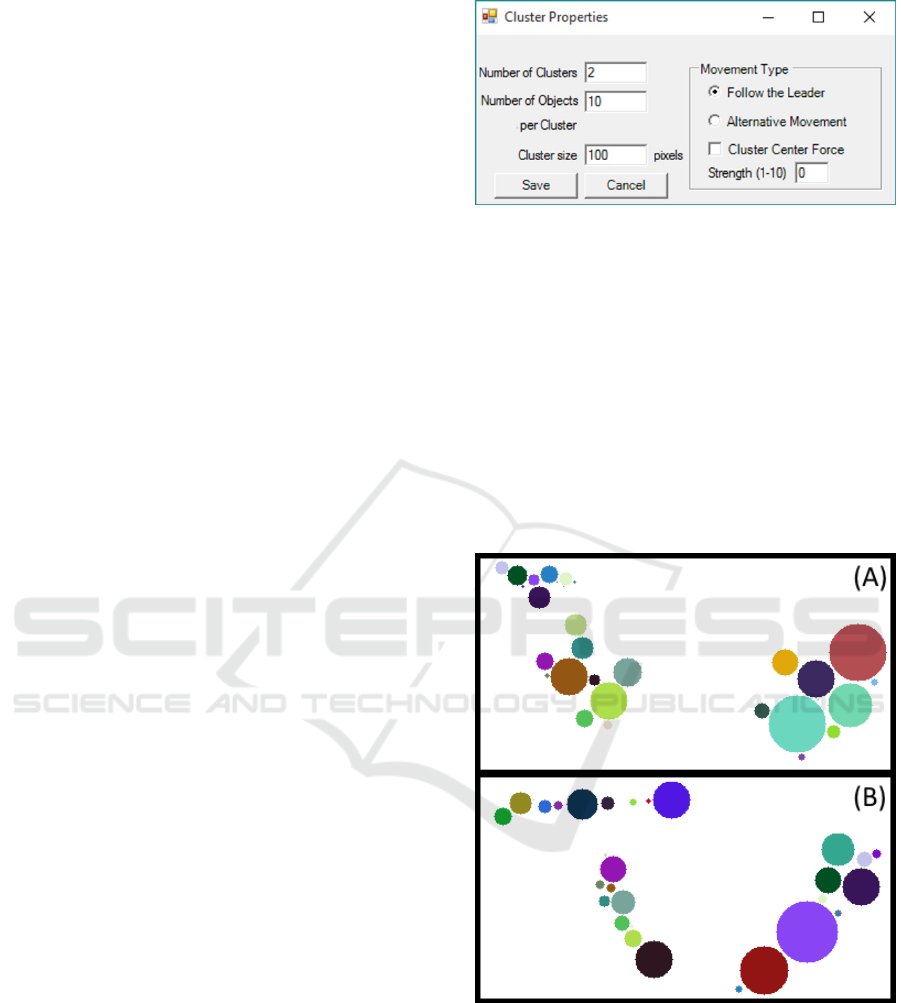
Properties” (shown in Figure 2) leads to a new
window with the options for clusters’ creation. Here,
one can choose the desired number of clusters, the
number of objects per cluster, and the size of the
clusters in pixels. It is also possible to choose
between two types of objects’ kinetics: “Follow the
Leader” and “Alternative Movement”. The
application of “Cluster Centre Force” and its
strength are shown in Figure 3 and explained in
Section 3.2.2.
3.2 Object Modelling
This sub-section focuses on the modelled features,
Figure 1: Image Generator Tool interface.
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
48
namely object movement and cluster, based on
biologically inspired objects, such as bacteria cells.
3.2.1 Object Motility
According to the user’s selection, objects can have
movement respecting a number of physical rules. If
this option is deactivated, objects will move
arbitrarily through the image.
Objects in each frame are only represented by
their position in coordinates in x and y and their
morphology factor. In each frame, each object can
move to a new x and y coordinates by an arbitrary
distance that cannot be higher than the “Maximum
Velocity” value in pixels.
With occlusions and “exits and entries” also
deactivated, objects will avoid the positions where
they collide with other objects or go out the image
boundaries, searching for a position considering
these limitations and the maximum distance they can
move between frames.
If the user chooses to give objects “Physical
Move”, in addition to previous features, each object
will have a velocity and an orientation (physical
parameters) assigned to it, meaning that their
position dynamics will depend on these two values.
In each frame, each object will have new x and y
coordinates distanced “d” (no bigger than
“Maximum Velocity”) from the previous frame
coordinates, direction “o” (between 0 and 2pi
radians), with both components using an
independent random variable.
If entries and exits are deactivated and if an
object is heading to the image boundary, it is
reflected respecting Snell’s Law, causing a change
in the angle’s direction of movement. If occlusions
are deactivated, when two objects are about to
collide, they change to opposite orientations in an
approximation to the reflection laws, but ignoring
differences in their morphologies.
3.2.2 Cluster Creation
When selecting the option “Create Clusters”, the
Generator will create a time-series with the number
of clusters, objects and size of cluster chosen by the
user.
These options (shown in Figure 2) must be
consistent and take into consideration the image
size.
In “Alternative Movement” (as shown in Figure
3-A) all objects of each cluster have the same
physical parameters, which means that they move in
the same direction with the same speed (with a small
independent arbitrary component).
Figure 2: Interface options for cluster properties.
In the “Follow the Leader” movement mode (as
shown in Figure 3-B), each cluster has a leading
object. The characteristics of the other objects of the
same cluster are dependent on the leader’s
behaviour. The leader “receives” the physical
parameters at first frame (velocity and orientation)
and at each frame the other objects of its cluster will
move in the leader’s direction, minimizing the
distance to it, but respecting the “non-collision” rule.
If two objects from different clusters collide, one of
them will start belonging to the other cluster. This
may cause the “merging” of clusters.
Figure 3: Exemplificative frames of (a) ‘Alternative’
Movement (b) ‘Follow the leader’ Movement.
The “Cluster Centre Force” feature is exclusively
for “Alternative Movement” that creates an
attraction force at the cluster’s centre, with a
selectable strength selected by the user. This force
keeps cluster’s objects together, even when colliding
with the image borders or other objects. Increasing
the strength, the objects will move faster to the
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
49
cluster’s centre. In this mode of motility, when
objects from different clusters collide, they will be
“left behind” by their cluster until they can join it
again.
3.3 Tested Tracking Algorithms
3.3.1 Simple Nearest-Neighbour Algorithm
The first tracking algorithm tested was the Simple
Nearest-Neighbour (NN). This method only takes
into consideration the position of each object in each
frame of the time-series, and uses the Euclidian
Distance between points to find matching objects
between frame n and n+1. Being
the distance
between two objects:
(1)
Where
and
are the positions of each object in
frame n and
and
are the positions in frame
n+1. Having the distance between each object in
frame n and all objects in frame n+1,
correspondences are made based on the minimum
distance. The object in frame n+1 closer to each
object in frame n is assigned to it. If two objects in
n+1 are assigned to the same object in n, the closer
object is assigned, until all correspondences between
frames are unique (Elfring et al., 2010).
3.3.2 Nearest-Neighbour with Morphology
Algorithm
The next algorithm tested was the Nearest
Neighbour with Morphology (NNm). This method
accounts not only for the differences between the
positions of each object in each frame, but also for a
shape-related factor, called morphology.
This algorithm calculates the distance percolated
by each object between frames n and n+1 using
equation (1). Being
the morphology of each
object in frame n, and
the shape factor in n+1,
the difference,
, between these variables is
calculated by:
|
|
(2)
The total difference,
, between each object in
each frame pair is given by (3) with and being
the weights given to each partial distance.
∙
∙
(3)
Here different weights are used (as presented in the
Results section), in order to study the best way to
combine them, to achieve the best possible results.
3.3.3 Cluster Tracking
Identifying clusters is one of the most complex
issues of image characterization (Czink et al., 2006).
In this work, the problem lays in tracking objects
knowing that they are grouped in clusters. Bacteria
often group in this way, so the goal is to find a
method that improves tracking of clustered objects.
One of the main problems of clustered objects is
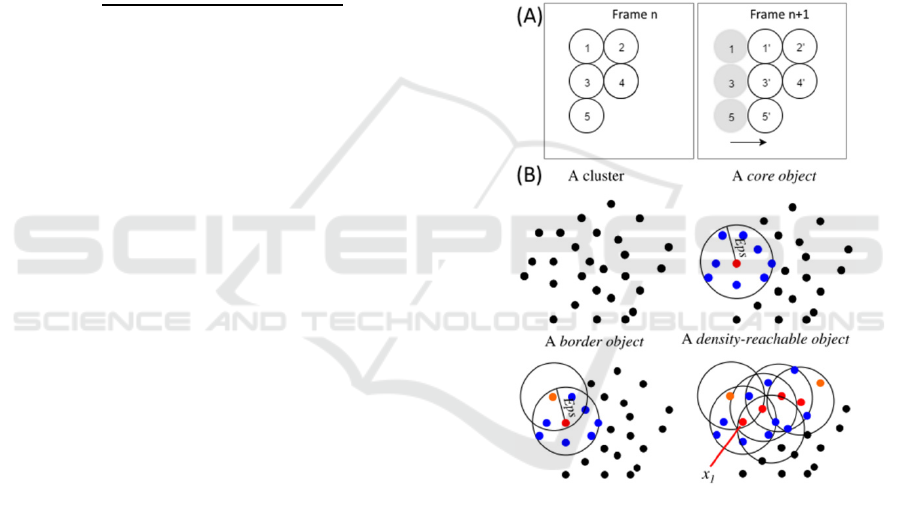
illustrated in Figure 4-A. Using Nearest-Neighbour
(or Nearest Neighbour with Morphology) to track
these frames, the algorithm will immediately
misidentify at least two of the objects of frame n+1.
This will occur in objects 1’ and 3’, and it happens
because their position in n+1 is exactly the same that
objects 2 and 4 have in n.
Figure 4: (A) Example of a possible misidentification
using the Nearest-Neighbour Algorithms. (B) After
defining ‘MinPts’ as the minimal number of objects in the
neighbourhood, and Eps as the neighbourhood radius, we
can define a core object (Red) when its local density is
higher than ‘MinPts’ and a border object (Orange) if its
local density is less than ‘MinPts’. Two density-reachable
objects are defined if there exists a chain of core objects
with distances between them smaller than Eps. Adapted
from (Tran et al., 2013).
To solve this problem we choose to implement a
novel tracking algorithm that considers the cluster’s
features and singularities. The developed method to
track clustered objects has several steps, and the first
is to correctly identify the clusters and objects
belonging to them. The method is called Density-
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
50

Based Spatial Clustering of Applications with Noise
(DBSCAN) (Ester et al., 1996) in its revised version
(Tran et al., 2013). This method formalizes the
notion of “cluster” and “noise”, using the definition
of density to characterize clusters, which means that
to define a cluster, the density of the neighbourhood
of each point has to be higher than a given threshold.
‘MinPts’ are the minimal number of objects in the
neighbourhood, and Eps is the neighbourhood radius
(see figure 4-B).
Objects can be divided in three categories: core,
border and noise (see figure 4-B). An object is a core
object if its local density is higher than ‘MinPts’. It
is considered a border object if its local density is
less than ‘MinPts’ and it belongs to the
neighbourhood of a core object. An object is
classified as a noise if in its Eps radius there are less
than ‘MinPts’ objects and none is a core.
Finally, we identify two density-reachable
objects if there exists a chain of core objects
between them (see Figure 4-B), with distances
between them smaller than ‘MinPts’ (Tran et al.,
2013).
This approach improves clustering identification
when data has dense adjacent clusters (Tran et al.,,
2013). They also introduced the core-density-
reachable objects, which are the same as the chain of
density-reachable objects, but cutting border objects
from chain’s ends and staying unclassified until all
core objects are identified (Tran et al., 2013).
The algorithm has two main steps: ‘dbscan’ and
‘ExpandCluster’. The first step lies in covering each
object and running ‘ExpandCluster’ if the object is
unclassified. Then, it returns all objects that are
core-density-reachable from that one. If it is a core
object, a cluster is produced. If it is a border object,
it has no core-density-reachable objects, and follows
to the next one. After all chains from the core object
are known, it is assigned to its best density-reachable
chain and all border objects.
After identifying the clusters in all frames with
DBSCAN, a novel algorithm for object tracking was
developed. This algorithm assumes that objects are
grouped and move in clusters, treating each cluster
as a separate individual while tracking. The first step
(with all clusters identified) is to isolate the clusters
and calculate their centroid, in coordinates x and y:
∑
(4)
After all centroids are calculated, they are processed
as objects, since they have their own coordinates.
The Nearest-Neighbour algorithm is then applied to
these coordinates, tracking the clusters and resulting
in a sequence of results similar to object tracking but
treating a cluster individually.
4 RESULTS AND DISCUSSION
We generated several time-series that can be used as
a benchmark to test tracking algorithms. For this, we
simulated examples with different starting number
of objects (20 to 160) and ‘Maximum Velocity’
(V=5, 10, 15, 20 and 30).
The generated images have a 1000x500 pixel
size (first and second experiment) and 1500x100
(third experiment). The implemented Tracking
Algorithms automatically processes the csv files
with the objects’ true positions produced by the
Image Generator. The detected object tracking is
then compared with the gold standard, where one
error is counted when one object is incorrectly
tracked from one frame to another it is considered a
False Positive (FP). When one object is tracked
correctly between two consecutive frames it is
considered a True Positive (TP). It is important to
notice that errors that occur in the beginning of the
time series are typically propagated through the
entire sequence. We present in the following tables
the tracking errors (false discovery rate), calculated
as FP/(FP+TP).
4.1 Simple Nearest-Neighbour
Algorithm
We tested 10 time-series of 100 frames for each
example with different objects and different
maximum velocity. In Table 1, we present the
tracking performance of the Simple Nearest-
Neighbour Algorithm, based on the ground-truth
produced by the image generator. A tracking error is
calculated on every frame and accumulated to the
end of the time-series.
Table 1: Tracking errors of the Simple Nearest-Neighbour
Algorithm.
Obj. V=5 V=10 V=15 V=20 V=30
20 0,00 0,92 1,06 4,19 19,20
40 0,26 1,27 3,23 5,93 24,01
60 0,06 1,58 5,63 12,38 39,66
80 0,24 1,84 6,62 15,74 45,06
100 0,27 1,20 7,85 19,94 49,76
120 0,22 1,69 10,57 21,16 51,86
140 0,55 3,71 14,16 26,57 58,07
160 0,42 4,12 14,91 33,74 63,89
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
51
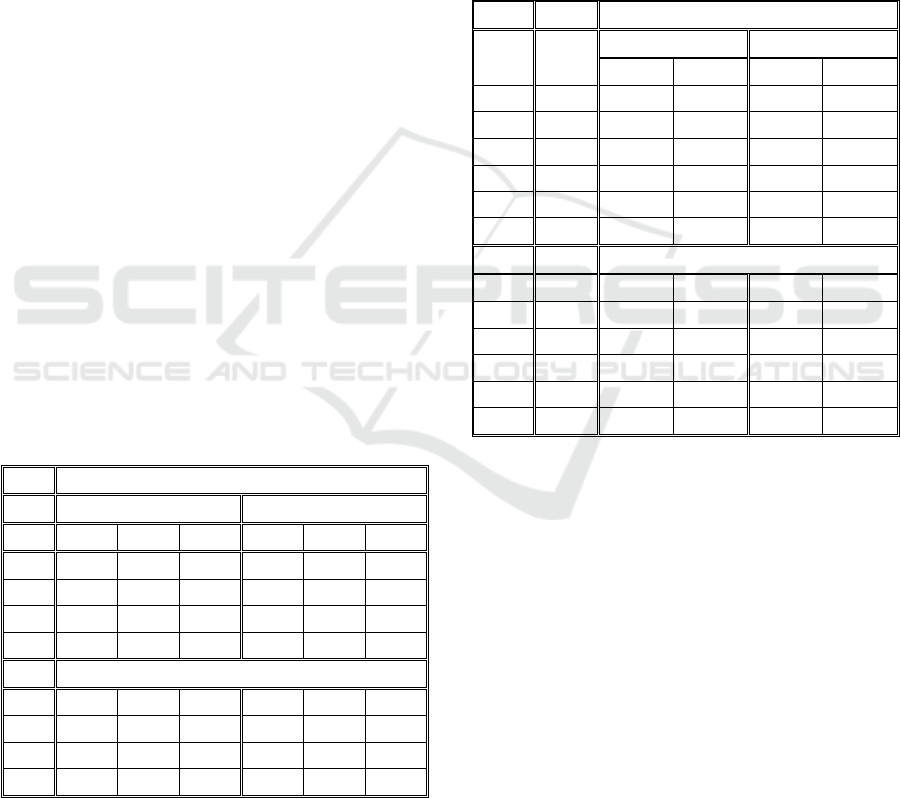
In this case, the morphology shape-related factor
called was set at 0.05 (this value was chosen to
emulate biologically inspired objects that slowly
change their shape over time).
The results from Table 1 show that this simple
algorithm can still handle the increase in the number
of objects while keeping a small velocity, and that
increasing the velocity from 15 to 20 and 30 the
tracking performance was significantly reduced.
4.2 Nearest-Neighbour with
Morphology Algorithm
In this second experiment, we show how tracking
taking into account the morphology of the object can
be helpful in the worst case scenario of the last
experiment. In Table 2, we present the results of the
tracking performance of the Nearest-Neighbour with
Morphology Algorithm.
In this case we also produced 10 time-series of
100 frames for each example with different objects
and different maximum velocity, but also with
distinct morphology factors.
We tested the algorithm in two configurations;
the first giving a 60% importance to the calculated
distance between objects ( factor in equation 3) and
40% to the calculated morphology difference (
factor). For the second configuration we used 40%
for and 60% for . Here we also changed the
shape-related factor and used both 0.05 and 0.1
values.
Table 2: Tracking errors of the Nearest-Neighbour with
Morphology Algorithm.
= 60% and =40%
m factor= 0.05 m factor= 0.1
Obj. V=15 V=20 V=30 V=15 V=20 V=30
100 6,02 14,71 41,60 6,03 17,79 40,55
120 6,27 14,92 42,05 9,24 19,68 44,45
140 9,29 18,34 48,96 8,34 21,59 48,90
160 10,32 25,49 55,35 10,26 25,39 55,32
Obj.
= 40% and =60%
100 4,26 10,57 33,37 3,96 12,07 32,44
120 4,53 10,80 33,95 6,39 14,07 37,09
140 6,36 14,58 39,30 5,34 15,18 41,36
160 7,27 20,78 46,81 8,06 18,93 49,38
From Table 2, we observe that tracking results
can be improved by using the Morphology
Algorithm (e.g. in the worst case scenario the error
percentage was reduced from 64% to 47%).
4.3 Cluster Tracking
The Create Clusters property was used to test the
same tracking algorithms (Simple NN and NN with
Morphology Algorithms with = 40%). The
simulated parameters were: number of clusters (1, 5
or 10), number of objects per cluster (10 or 15),
maximum velocity (5 or 10), Alternative Movement,
Center Force (4) and morphology factor (0 or 0.05).
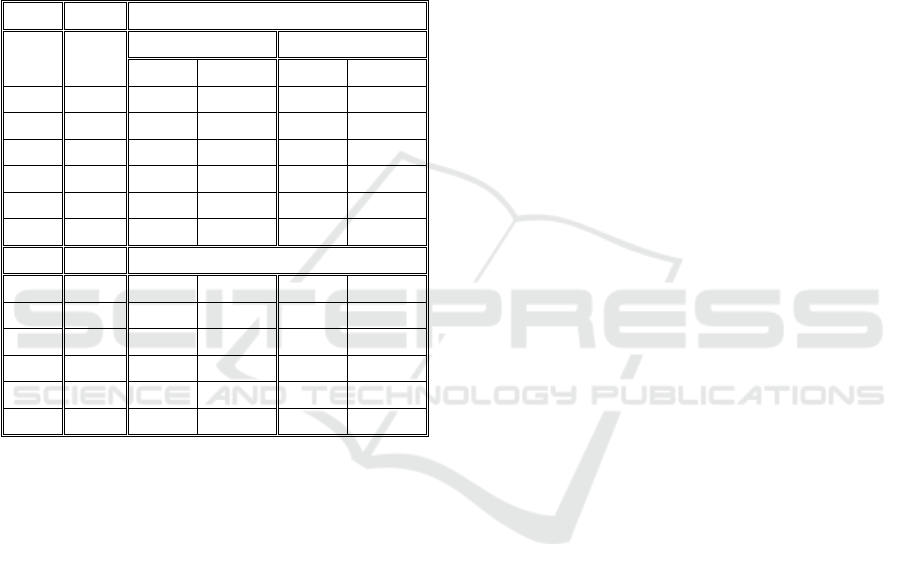
The tracking results are presented in table 3.
Table 3: Tracking errors, within clusters with different
properties, using the Simple and Morphology Nearest-
Neighbour Algorithms.
Simple NN Algorithm
Nº of
Clust
Obj. /
Clust.
m factor= 0 m factor= 0.05
V=5 V=10 V=5 V=10
1 10 7,79 30,42 9,88 23,33
1 15 11,74 50,91 10,74 38,06
5 10 7,48 34,71 10,95 31,89
5 15 17,43 45,22 16,06 44,51
10 10 12,20 38,26 11,64 42,47
10 15 21,14 53,90 23,52 57,34
NN with Morphology
1 10 1,27 4,879 5,52 13,83
1 15 3,76 21,15 4,63 20,76
5 10 1,80 12,98 4,69 15,93
5 15 7,16 20,77 5,94 22,07
10 10 3,78 16,15 4,55 19,71
10 15 8,73 28,36 10,13 34,12
For the Cluster creation, we used 10 time-series
(and averaged the results) of 200 frames and
calculated the object tracking on every frame and
accumulated to the end of the time-series.
The DBSCAN algorithm tries to separate each
cluster in every frame. Therefore, if the number of
clusters is the same between the actual frame and the
previous one (t and t-1), then they are matched using
NN, treating them as isolated objects and aligned
using their centroids. If the number of clusters
changes, we skip the first step and check the number
of objects inside each cluster. When there are more
objects in t then in t-1, these ‘extra’ objects are
called ‘Possible Entry’, if there are less objects, they
are called ‘Possible Exit’.
This classification is done temporarily and
compares the "Possible Exit" features to the features
of all other objects of the frame t-1, linking it to a
"Possible entry" in another cluster (meaning that it
left one cluster to join another), classifying it as
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
52
noise, or as an object leaving the image. The main
difference between the two DBSCAN Algorithms is
that, in the first, this classification is done after the
tracking and in the second it is done before the
tracking, equalizing the number of objects between
the clusters.
Results from both DBSCAN Algorithms are
presented in Table 4. We can see that DBSCAN
Algorithms do not improve significantly over the
Nearest-Neighbour with Morphology.
Table 4: Tracking errors, within clusters with different
properties, using two different DBSCAN Algorithms.
DBSCAN 1
Nº of
Clust
Obj. /
Clust.
m factor= 0 m factor= 0.05
V=5 V=10 V=5 V=10
1 10 5,55 2,97 9,64 9,14
1 15 1,92 12,94 3,87 10,49
5 10 5,43 13,71 6,42 16,61
5 15 5,84 19,29 6,49 20,84
10 10 5,81 17,55 6,42 21,07
10 15 8,65 28,29 9,91 34,52
DBSCAN 2
1 10 4,67 2,97 9,64 7,82
1 15 2,59 12,96 3,87 10,49
5 10 5,54 14,56 7,55 17,37
5 15 5,74 19,35 6,19 20,82
10 10 6,00 17,57 6,43 21,42
10 15 8,65 28,29 9,98 34,52
A strange behaviour with just 1 cluster was
identified in the DBSCAN algorithms, where
increasing the objects actually decreased the
tracking errors. This behaviour could be due to a
bigger movement restriction within clusters with
higher objects, but further studies are required to
analyse this behaviour.
5 CONCLUSIONS AND FUTURE
WORK
To support high-throughput experiments of single
cell imaging, reliable automated image processing
methods are required. Although most studies focus
on automatic segmentation of cells or cellular
structures, in a time-series a proper object tracking is
also necessary, particularly since errors in tracking
are propagated, meaning that even small tracking
errors (particularly on the initial frames) can lead to
a high percentage of misidentified tracks.
To validate such Tracking Algorithms, it is
necessary to use a labelled ‘ground truth’.
Sometimes this ground-truth is manually processed,
which can be unfeasible in a Big Data scenario. A
more viable alternative is to generate artificial
images by simulating biological cell models. To
produce such artificial images we developed an open
source platform that can simulate biologically
inspired bacterial systems, by creating cells with
different morphologies, physical movement and
cluster creation. Using this Platform, we evaluated
three tracking algorithms (Simple Nearest-
Neighbour, Nearest-Neighbour with Morphology
and two variations of the DBSCAN Algorithm).
The obtained results showed that, for cases with
lower maximum velocity, the Simple Nearest-
Neighbour Algorithm was able to track objects even
with a significant increase in the number of objects.
The Nearest-Neighbour with Morphology algorithm
can help in reducing tracking errors when the
velocity is increased. In the example where we
forced the creation of clusters, both the Nearest-
Neighbour with Morphology and the DBSCAN
algorithms showed similar results. In the near future,
we plan to study and compare other tracking
methodologies in different cluster configurations.
We expect this open-sourced tool (available at:
http://griduni.uninova.pt/Clustergen/
ClusterGen_v1.0.zip) to help future endeavours in
the development of new tracking algorithms, as it
can produce huge amount of benchmarked data in
various configurations.
Future developments of this tool involve adding
an object division module, which can be used to test
division tracking in dense clusters. We also plan to
add a module that introduces secondary bodies
inside the primary objects, simulating internal cell
organelles and structures. A future application will
also be made available as a web-based system to
improve usability and compatibility.
ACKNOWLEGMENTS
Work supported by the Portuguese Foundation for
Science and Technology (FCT/MCTES) through a
PhD Scholarship, ref. SFRH/BD/88987/2012 to LM,
SADAC project (ref. PTDC/BBB-MET/1084/2012)
and by FCT Strategic Program UID/EEA/00066/203
of UNINOVA, CTS. This work is also funded by the
Academy of Finland [ref. 126803 to ASR].
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
53

REFERENCES
Bhattacharyya, A., 1943. On a Measure of Divergence
Between Two Statistical Populations Defined by
Probability Distributions. Bulletin of the Calcutta
Mathematical Society, 35, pp.99–110.
Bonnet, N., 2004. Some trends in microscope image
processing. Micron (Oxford, England : 1993), 35(8),
pp.635–653.
Coelho, L.P., Shariff, A. & Murphy, R.F., 2009. Nuclear
Segmentation In Microscope Cell Images A Hand-
Segmented Dataset And Comparison Of Algorithms.
In Proc IEEE Int Symp Biomed Imaging. pp. 518–521.
Coutu, D.L. & Schroeder, T., 2013. Probing cellular
processes by long-term live imaging--historic
problems and current solutions. Journal of cell
science, 126(Pt 17), pp.3805–15.
Czink, N., Mecklenbräuker, C. & Del Galdo, G., 2006. A
novel automatic cluster tracking algorithm. IEEE
International Symposium on Personal, Indoor and
Mobile Radio Communications, PIMRC, pp.1–5.
Danuser, G., 2011. Computer vision in cell biology. Cell,
147(5), pp.973–8.
Deshmukh, M. & Bhosle, U., 2011. A survey of image
registration. International Journal of Image
Processing, 5(3), pp.245–269.
Elfring, J., Janssen, R. & van de Molengraft, R., 2010.
Data Association and Tracking: A Literature Survey.
In ICT Call 4 RoboEarth Project.
Ester, M. et al., 1996. A Density-Based Algorithm for
Discovering Clusters in Large Spatial Databases with
Noise. In 2nd Int. Conference on Knowledge
Discovery and Data Mining. pp. 226–231.
Frigault, M. et al., 2009. Live-cell microscopy - tips and
tools. Journal of Cell Science, 122(6), pp.753–767.
Gorji, A. & Menhaj, M.B., 2007. Multiple Target
Tracking for Mobile Robots Using the JPDAF
Algorithm. In 19th IEEE International Conference on
Tools with Artificial Intelligence (ICTAI 2007). pp.
137–145.
Gotelli, N.J. & McGill, B.J., 1996. Null versus neutral
models: what’s the difference? Ecography, 29(5),
pp.793–800.
Gu, S., Zheng, Y. & Tomasi, C., 2011. Efficient visual
object tracking with online nearest neighbor classifier.
In Computer Vision – ACCV 2010. Volume 6492 of the
series LNCS. pp. 271–282.
Häkkinen, A. et al., 2013. CellAging: a tool to study
segregation and partitioning in division in cell lineages
of Escherichia coli. Bioinformatics (Oxford, England),
29(13), pp.1708–1709.
Joyce, J., 2014. Kullback-Leibler Divergence. In M.
Lovric, ed. International Encyclopedia of Statistical
Science SE - 327. Springer Berlin Heidelberg, pp.
720–722.
Kruse, K., 2012. Bacterial Organization in Space and
Time. In Comprehensive Biophysics. pp. 208–221.
Lehmussola, A. et al., 2007. Computational framework for
simulating fluorescence microscope images with cell
populations. IEEE transactions on medical imaging,
26(7), pp.1010–6.
Lehmussola, A. et al., 2011. Synthetic Images of High-
Throughput Microscopy for Validation of Image
Analysis Methods. Proceedings of the IEEE, 96(8),
pp.1348 – 1360.
Martins, L., Fonseca, J. & Ribeiro, A., 2015. “miSimBa” -
A simulator of synthetic time-lapsed microscopy
images of bacterial cells. In Proceedings - 2015 IEEE
4th Portuguese Meeting on Bioengineering, ENBENG
2015. pp. 1 – 6.
Meijering, E., 2012. Cell Segmentation: 50 Years Down
the Road. IEEE Signal Processing Magazine, 29(5),
pp.140–145.
Misteli, T., 2007. Beyond the sequence: cellular
organization of genome function. Cell, 128(4),
pp.787–800.
Murphy, R., 2012. CellOrganizer: Image-derived Models
of Subcellular Organization and Protein Distribution.
Methods in Cell Biology, 110, pp.179–93.
Ruusuvuori, P. et al., 2008. Benchmark Set Of Synthetic
Images For Validating Cell Image Analysis
Algorithms. In Proceedings of the 16th European
Signal Processing Conference, EUSIPCO.
Satwik, R. et al., 2012. SimuCell : a flexible framework
for creating synthetic microscopy images a
PhenoRipper : software for rapidly profiling
microscopy images. , 9(7), pp.634–636.
Selinummi, J. et al., 2005. Software for quantification of
labeled bacteria from digital microscope images by
automated image analysis. BioTechniques, 39(6),
pp.859–863.
Shi, J. & Tomasi, C., 1994. Good features to track. In
Proceedings CVPR’94. 1994 IEEE Computer Society
Conference on. IEEE. pp. 593–600.
Sliusarenko, O. & Heinritz, J., 2011. High-throughput,
subpixel precision analysis of bacterial morphogenesis
and intracellular spatio-temporal dynamics. Molecular
Microbiology, 80(3), pp.612–627.
Smith, A., 1993. Novel approach to nonlinear/non-
Gaussian Bayesian state estimation. Radar and Signal
Processing, IEE Proceedings F, 140(2), pp.107–113.
Sung, M.-H. & McNally, J.G., 2011. Live cell imaging
and systems biology. Wiley interdisciplinary reviews.
Systems biology and medicine, 3(2), pp.167–82.
Svoboda, D. et al., 2007. On simulating 3D Fluorescent
Microscope Images. In Computer Analysis of Images
and Patterns -12th International Conference, CAIP
2007, Vienna, Austria, August 27-29, 2007.
Proceedings. pp. 309–316.
Svoboda, D., Kozubek, M. & Stejskal, S., 2009.
Generation of digital phantoms of cell nuclei and
simulation of image formation in 3D image cytometry.
Cytometry. Part A : the journal of the International
Society for Analytical Cytology, 75(6), pp.494–509.
Tissainayagam, P. & Suter, D., 2005. Object tracking in
image sequences using point features. Pattern
Recognition
, 38(1), pp.105–113.
Tran, T.N., Drab, K. & Daszykowski, M., 2013. Revised
DBSCAN algorithm to cluster data with dense
SIMULTECH 2016 - 6th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
54

adjacent clusters. Chemometrics and Intelligent
Laboratory Systems, 120, pp.92–96.
Ulman, V., Oremus, Z. & Svoboda, D., 2015. TRAgen: A
Tool for Generation of Synthetic Time-Lapse Image
Sequences of Living Cells. In Proceedings of 18th
International Conference on Image Analysis and
Processing (ICIAP 2015). Springer International
Publishing, pp. 623–634.
Wang, Q. et al., 2010. Image segmentation and dynamic
lineage analysis in single-cell fluorescence
microscopy. Cytometry A, 77(1), pp.101–110.
Wyawahare, M., Patil, P. & Abhyankar, H., 2009. Image
Registration Techniques : An overview. Int. Journal of
Signal Processing, Image Processing and Pattern
Recognition, 2(3), pp.11–28.
Xiong, W. et al., 2010. Learning Cell Geometry Models
For Cell Image Simulation : An Unbiased Approach.
In Proceedings of 2010 IEEE 17th International
Conference on Image Processing. pp. 1897–1900.
Yilmaz, A., Javed, O. & Shah, M., 2006. Object tracking:
A survey. ACM Computing Surveys, 38(4, Article 13),
pp.1–45.
Young, J. et al., 2012. Measuring single-cell gene
expression dynamics in bacteria using fluorescence
time-lapse microscopy. Nat. Protoc., 7(1), pp.80–8.
Zhao, T. & Murphy, R.F., 2007. Automated learning of
generative models for subcellular location: building
blocks for systems biology. Cytometry. Part A : the
journal of the International Society for Analytical
Cytology, 71(12), pp.978–90.
Zhou, H., Yuan, Y. & Shi, C., 2009. Object tracking using
SIFT features and mean shift. Computer Vision and
Image Understanding, 113(3), pp.345–352.
An Image Generator Platform to Improve Cell Tracking Algorithms - Simulation of Objects of Various Morphologies, Kinetics and
Clustering
55
