Input Encoding Proposal for Behavioral Experiments with a Virtual
C. elegans Representation
Gorka Epelde
1
, Andoni Mujika
1
, Roberto Álvarez
1
, Peter Leškovský
1
, Alessandro de Mauro
1
,
Finn Krewer
2
and Axel Blau
3
1
eHealth and Biomedical, Vicomtech-IK4, Mikeletegi Pasealekua, Donostia-San Sebastián, Spain
2
College of Engineering and Informatics, National University of Ireland Galway, University Road, Galway, Ireland
3
Dept. of Neuroscience and Brain Technologies (NBT), Fondazione Istituto Italiano di Tecnologia (IIT), 16163 Genoa, Italy
Keywords: Behavioral Experiment Input Encoding, C, elegans, Complex Systems Simulation, in Silico Simulation.
Abstract: This paper discusses a Caenorhabditis elegans (C. elegans) nematode behavioral experiment input
encoding. It proposes a common digital representation for behavioral studies with C. elegans. This work is a
step forward towards the reproducibility and comparability of in silico simulations of the nematode with
real-world experiments. The digital representation is divided into environmental and experimental
configurations. The behavioral input is structured by duration-based behavioral experiment types at the top
level (i.e. interaction at a specific time, interaction from t
0
– t
1
and overall duration) and by interaction type
(i.e. mechanotaxis, chemotaxis, thermotaxis, galvanotaxis and phototaxis) for each duration-based type. The
environment configuration is composed of the identification of the worm’s mutation type, worm crowding,
initial location, configuration of the assay plate, and obstacle settings. Parameters are defined by an XML
schema to ensure the interoperability with other simulation solutions. It is being implemented and tested in
the context of the Si elegans project.
1 INTRODUCTION
The reproducibility and comparability of
experiments is key to scientific progress. Small
variations in the conditions of an assay or in the
applied behavioral stimuli can dramatically change
the results of an experiment (Hart, 2005). Based on
this consideration, researchers usually detail their in
vivo experiments with respect to the animal’s
properties, the environmental conditions and the
applied stimuli.
For the animal of interest, the nematode
Caenorhabditis elegans (C. elegans), the scientific
community has already detailed the requirements for
in vivo behavioral assays such as the definition of
the type of animals to be assayed with respect to
control strains, their feeding status, cultivation
conditions, the ambient conditions, the scoring of
perceived behavior, the statistical analysis and the
reporting of results (Hart, 2005). Once defined, the
different assay considerations detail each C. elegans
behavioral experiment, which can then be grouped
by the observed behavioral response types.
With the advent of the in silico modelling and
simulation of living organisms in different
biomedical research areas (e.g., computational
neuroscience or systems biology), the minimum
information content of a simulation needs to be
agreed on in order to ensure reproducibility within
simulations, comparability between different
simulation systems and comparability between in
vivo and in silico experiments (Waltemath et al.,
2011). In this respect, different initiatives have
attempted to standardize the computational models,
simulation experiment definitions, graphical
visualizations, results provision, and vocabularies
including the provision of guidelines on their
effective reuse. To the best of our knowledge the
existing approaches do not support the encoding of
inputs for behavioral experiments with simple life
forms but rather target on the description of inputs at
signal level, as is considered by distinct biological
models of cells or neurons.
The contribution of this paper is the proposal of
a digital representation to encode simulated
experiments and stimuli at behavioral level for C.
elegans.
Epelde, G., Mujika, A., Alvarez, R., Leškovský, P., Mauro, A., Krewer, F. and Blau, A..
Input Encoding Proposal for Behavioral Experiments with a Virtual C. elegans Representation.
In Proceedings of the 3rd International Congress on Neurotechnology, Electronics and Informatics (NEUROTECHNIX 2015), pages 115-120
ISBN: 978-989-758-161-8
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
115
We first analyze the related work in the field of
simulation experiment reproducibility (Section 2).
Section 3 presents an innovative approach to define
the behavioral experiment input for the C. elegans
nematode simulation experiments. In Section 4, we
summarize the paper and present our conclusions.
2 RELATED WORK
In the in silico biomedical modelling and simulation
research areas, efforts have aimed at standardizing
different models and processes as well as integrating
specialized software into unified platforms (Amari et
al., 2002; Cannon et al., 2007; Teeters et al., 2008;
Ghosh et al., 2011; Dräger et al., 2014). Their main
goal was the reproducibility, comparability of
simulated experiments and the reusability of
building blocks to provide the integration of more
complex simulations or of multilevel modelling and
simulation tools.
In the systems biology research area, (Dräger et
al., 2014) examined diverse modelling standards and
data formats that are currently in use within the
scientific community together with databases, from
which relevant resources that conform with these
formats can be obtained. (Ghosh et al., 2011)
described the types of software tools that are
required at different research stages, current options
that are available for researchers, challenges and
prospects for modelling the effects of genetic
changes on physiology and the concept of an
integrated biomedical research platform.
In the computational neuroscience research area,
(Cannon et al., 2007; Davison et al., 2009; Crook et
al., 2013) examined the interoperability and the
interfacing of neuroscience modelling software and
neuronal network simulations. (Amari et al., 2002;
Teeters et al., 2008) discussed the data sharing and
the integration of shared databases and tools for
integrative neuroscientific research.
(Dräger et al., 2014) suggested a structure to
revise the state of the art in standardization and
interoperability efforts. The review is structured into
modeling guidelines, model encoding formats,
simulation procedures, graphical model visualization
and numerical results representation.
Modelling guidelines generally include
requirements for the minimum information given in
an experiment as well as ontologies that describe the
controlled term vocabularies that should be used in
the model encoding formats. The Minimum
Information Required in the Annotation of Models
(MIRIAM) guidelines promote the exchange and
reuse of biochemical computational models (Le
Novère et al., 2005). While mentioning the need for
result reproducibility, MIRIAM does not cover the
minimum requirements necessary for simulating the
models. In contrast, the Minimum Information
About a Simulation Experiment (MIASE) sets out to
define the minimum requirements for simulation
descriptions, and allows thus for unambiguous
reproduction of experiment (Waltemath, Adams,
Beard, et al., 2011). With respect to ontologies, the
Kinetic Simulation Algorithm Ontology (KiSAO)
organizes algorithms to simulate models (Courtot et
al., 2014), and the Systems Biology Ontology (SBO)
proposes a collection of terms that describe the
structure of a model, its components as well as
modeling frameworks and processes (Courtot et al.,
2014). With a focus on C. elegans, a hierarchically
structured, controlled vocabulary of terms that
standardizes phenotype descriptions, namely the
Worm Phenotype Ontology (WPO), is defined
(Schindelman et al., 2011).
Several formats that encode biological models
have been proposed. The most representative
formats include the Systems Biology Markup
Language (SBML) to represent biochemical network
models (Hucka et al., 2003), CellML for defining
mathematical models of cellular functions (Lloyd et
al., 2004) and NeuroML for describing data-driven
models of neurons and neural networks (Gleeson et
al., 2010). Regarding inter-model interoperability,
PyNN (Python Neural Networks) is a programming
interface common to multiple neuronal network
simulators, which allows to write a simulation script
once in the Python programming language and run it
without modification on any supported simulator
(Davison et al., 2009).
With respect to simulation procedures, the
Simulation Experiment Description Mark-up
Language (SED-ML) provides a standardized,
machine-readable format (Waltemath, Adams,
Bergmann, et al., 2011) for the information required
by MIASE to enable the reproduction of simulation
experiments (Waltemath, Adams, Beard, et al.,
2011). Besides models identification, MIASE
requires a precise description of simulation steps
together with all the information for obtaining
numerical results to be reported in scientific
publications. In SED-ML documents, the simulation
experiment input is defined at a low model
parametric level because the models used e.g.,
SBML, CellML or NeuroML are biological models.
It is therefore not well suited for defining the
behavioral experiment input of a living organism in
an in silico simulation.
NeBICA 2015 - Symposium on Neuro-Bio-Inspired Computation and Architectures
116
Graphical visualizations of biological models
have been used to facilitate the understanding of a
model in publications and textbooks. As in other
analyzed parts of a simulation component,
standardization efforts have led to visualization
recommendations and data formats for the exchange
of the resulting visualization. Representative efforts
are (i) the Systems Biology Graphical Notation
(SBGN) (Le Novère et al., 2009) and the
corresponding markup language (SBGN-ML)
developed to visualize, store and exchange the
visualization of biological networks (Iersel et al.,
2012); and (ii) a framework defined for visualization
of CellML models, which allows the visualization of
the physical model or its biological interpretation
(Wimalaratne et al., 2009).
Finally, specific file formats have been proposed
for numerical results provision. The Numerical
Markup Language (NuML) (NuML Project, 2015),
originates from the numerical aspects of the Systems
Biology Results Markup Language (SBRML) (Dada
et al., 2010), with the aim of reusing it in multiple
other standardization efforts. In the area of
presenting results, ongoing work is carried out by
the OpenWorm project to validate the results of
behavioral simulation experiments and to compare
the perceived locomotion behavior of in vivo
experiments with in silico experiments (OpenWorm
Project, 2015).
Behavioral experiments are run by psychologists
for neurobehavioral research on humans (Mueller et
al., 2014) by using tools such as (Neurobehavioral
Systems, 2015). But we do not see a direct way for
exploiting these approaches in living organisms’ in
silico experiments.
After reviewing the related work on
interoperability and standardization of
computational neuroscience and systems biology
research areas, no specific solution seems to exist
that tackles the behavioral experiment input
encoding in general or specific to the C. elegans
nematode.
3 MATERIAL AND METHODS
According to rule 2B of the MIESE guidelines
(Waltemath et al., 2011), all information needed for
the correct implementation of the necessary
simulation steps must be included through precise
descriptions or references to unambiguous
information sources. When it comes to projects such
as OpenWorm (Szigeti et al., 2014) or Si elegans
(Blau et al., 2014) that aim to simulate a complete
life form, a behavioral input encoding is required.
As an effort to standardize the C. elegans behavioral
experiments input encoding, the following proposal
is presented. It was inspired by a collection of C.
elegans behavior experiments described in the
WormBook (Hart, 2005) and in recent C. elegans in
vivo research reports (Gabel et al., 2007; Ward et al.,
2008). The latter cover behavioral responses to
behavioral input types that are not yet included in
the WormBook.
The proposed digital representation is divided in
the environment and the experiment configurations.
The behavioral experiment is structured by duration-
based behavioral experiment types at the top level.
The following three behavioral experiment types
have been defined:
▪ Interaction at a specific time t
▪ Interaction from t
0
to t
1
▪ Experiment-wide configuration
Each duration-based experiment allows for the
definition of one or more interactions of each type.
For each duration-based experiment type, each of
the behavioral input types (i.e. mechanotaxis,
chemotaxis, thermotaxis, galvanotaxis, and
phototaxis) as identified by (Hart, 2005; Gabel et al.,
2007; Ward et al., 2008) is allowed. Behaviors
reported by (Hart, 2005), which are not sensory
behavioral inputs (i.e. locomotion, feeding, egg-
laying, mating, reproduction or defecation), have not
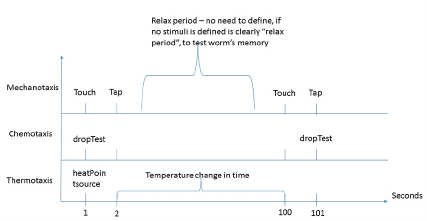
been considered. Figure 1 depicts an example of a
duration-based behavioral experiment.
Figure 1: Example of an experiment definition based on
different durations. It includes two temporal types:
“Interaction from t
0
– t
1
” for a change in temperature
labelled “Temperature change in time” and others of the
type “Interaction at specific time”.
On a third level, the concrete behavioral
experiments inputs are defined in the XML schema
developed by (Si elegans Consortium, 2015a).
An example experiment definition section of a
behavioral experiment input that contains multiple
interaction instances is presented in Figure 2.
Input Encoding Proposal for Behavioral Experiments with a Virtual C. elegans Representation
117

Figure 2: Example of the experiment definition section of
a behavioral experiment input encoding.
Figure 3: Example of the environment definition section of
a behavioral experiment input encoding.
The environment configuration structure and
elements have been extracted from the WormBook
experiments review (Hart, 2005). The environment
configuration is composed of the worm’s mutation
identification, crowding, location, plate
configuration, and obstacles configuration, which
have been parameterized. An example environment
definition section of a behavioral experiment input
that describes the experiment’s environment together
with the worm’s details is presented in Figure 3. A
complete behavioral experiment input encoding
example is provided in
(Si elegans Consortium,
2015b).
4 CONCLUSIONS AND FUTURE
WORK
In this paper, we have presented a proposal for a C.
elegans behavioral experiments input encoding
strategy. Its aim is to standardize the behavioral
input configuration and the simulation parameters
(e.g., environment configuration) for different C.
elegans nematode simulation frameworks. This
standardization effort targets at an interoperability
between these frameworks to allow for reproducible
and comparable simulations and for sharing
simulation building blocks. A brief review of the
state of the art of standardization and interoperability
efforts in the computational neuroscience and
systems biology research areas has led us to define a
behavioral experiment input encoding language from
scratch. The definition procedure was introduced
and the logic behind the markup language was
explained. Additionally, an example for an
experiment was provided together with the
corresponding XML schema of the defined markup
language; a complete example as part of the Si
elegans project simulation solution is available at the
following online repository: https://github.com/Si-
elegans/behavioural_experiment_definition . Future
work will include the development of import/export
modules for interoperability with similar simulation
platforms.
ACKNOWLEDGEMENTS
The Si elegans project is funded by the 7th
Framework Programme (FP7) of the European
Union under FET Proactive, call ICT-2011.9.11:
Neuro-Bio-Inspired Systems (NBIS). Finn Krewer is
supported by the Irish Research Council under the
EMBARK funding scheme.
NeBICA 2015 - Symposium on Neuro-Bio-Inspired Computation and Architectures
118
REFERENCES
Amari, Shun-Ichi, Beltrame, Francesco, Bjaalie, Jan G.,
Dalkara, Turgay, De Schutter, Erik, Egan, Gary F.,
Goddard, Nigel H., Gonzalez, Carmen, Grillner, Sten,
Herz, Andreas, Hoffmann, K.-Peter, Jaaskelainen, Iiro,
Koslow, Stephen H., Lee, Soo-Young, Matthiessen,
Line, Miller, Perry L., Da Silva, Fernando Mira,
Novak, Mirko, Ravindranath, Viji, Ritz, Raphael,
Ruotsalainen, Ulla, Sebestra, Vaclav, Subramaniam,
Shankar, Tang, Yiyuan, Toga, Arthur W., Usui, Shiro,
Van Pelt, Jaap, Verschure, Paul, Willshaw, David and
Wrobel, Andrzej (2002), Neuroinformatics: The
Integration of Shared Databases and Tools towards
Integrative Neuroscience. Journal of Integrative
Neuroscience, 01(02): 117–128.
Blau, Axel, Callaly, Frank, Cawley, Seamus, Coffey,
Aedan, Mauro, Alessandro De, Epelde, Gorka,
Ferrara, Lorenzo, Krewer, Finn, Liberale, Carlo,
Machado, Pedro, Maclair, Gregory, McGinnity,
Thomas-Martin, Morgan, Fearghal, Mujika, Andoni,
Petrushin, Alessandro, Robin, Gautier and Wade, John
(2014), The Si Elegans Project – The Challenges and
Prospects of Emulating Caenorhabditis Elegans, 436–
438, in: Duff, A., Lepora, N. F., Mura, A., Prescott, T.
J., and Verschure, P. F. M. J. (Eds.), Biomimetic and
Biohybrid Systems. Lecture Notes in Computer
Science. Springer International Publishing. available at
http://link.springer.com/chapter/10.1007/978-3-319-
09435-9_54 [2 October 2014].
Cannon, Robert C., Gewaltig, Marc-Oliver, Gleeson,
Padraig, Bhalla, Upinder S., Cornelis, Hugo, Hines,
Michael L., Howell, Fredrick W., Muller, Eilif, Stiles,
Joel R., Wils, Stefan and Schutter, Erik De (2007),
Interoperability of Neuroscience Modeling Software:
Current Status and Future Directions.
Neuroinformatics, 5(2): 127–138.
Courtot, M., Juty, N., Knupfer, C., Waltemath, D.,
Zhukova, A., Drager, A., Dumontier, M., Finney, A.,
Golebiewski, M., Hastings, J., Hoops, S., Keating, S.,
Kell, D. B., Kerrien, S., Lawson, J., Lister, A., Lu, J.,
Machne, R., Mendes, P., Pocock, M., Rodriguez, N.,
Villeger, A., Wilkinson, D. J., Wimalaratne, S., Laibe,
C., Hucka, M. and Le Novere, N. (2014), Controlled
Vocabularies and Semantics in Systems Biology.
Molecular Systems Biology, 7(1): 543–543.
Crook, Sharon M., Davison, Andrew P. and Plesser, Hans
E. (2013), Learning from the Past: Approaches for
Reproducibility in Computational Neuroscience, 73–
102, in: Bower, J. M. (Ed.), 20 Years of
Computational Neuroscience. Springer Series in
Computational Neuroscience. Springer New York.
available at http://link.springer.com/chapter/10.1007/
978-1-4614-1424-7_4 [23 September 2015].
Dada, Joseph O., Spasić, Irena, Paton, Norman W. and
Mendes, Pedro (2010), SBRML: A Markup Language
for Associating Systems Biology Data with Models.
Bioinformatics, 26(7): 932–938.
Davison, Andrew P., Brüderle, Daniel, Eppler, Jochen,
Kremkow, Jens, Muller, Eilif, Pecevski, Dejan,
Perrinet, Laurent and Yger, Pierre (2009), PyNN: A
Common Interface for Neuronal Network Simulators.
Frontiers in Neuroinformatics, 2. available at
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC26345
33/ [23 September 2015].
Dräger, Andreas and Palsson, Bernhard Ø. (2014),
Improving Collaboration by Standardization Efforts in
Systems Biology. Frontiers in Bioengineering and
Biotechnology, 2. available at http://www.ncbi.nlm
nih.gov/pmc/articles/PMC4259112/ [22 September
2015].
Gabel, Christopher V., Gabel, Harrison, Pavlichin, Dmitri,
Kao, Albert, Clark, Damon A. and Samuel,
Aravinthan D. T. (2007), Neural Circuits Mediate
Electrosensory Behavior in Caenorhabditis Elegans.
The Journal of Neuroscience, 27(28): 7586–7596.
Ghosh, Samik, Matsuoka, Yukiko, Asai, Yoshiyuki, Hsin,
Kun-Yi and Kitano, Hiroaki (2011), Software for
Systems Biology: From Tools to Integrated Platforms.
Nature Reviews Genetics. available at
http://www.nature.com/doifinder/10.1038/nrg3096 [22
September 2015].
Gleeson, P, Crook, S, Cannon, RC, Hines, ML, Billings,
GO, Farinella, M, Morse, TM, Davison, AP, Ray, S
and Bhalla, US (2010), NeuroML: A Language for
Describing Data Driven Models of Neurons and
Networks with a High Degree of Biological Detail.
PLoS Computational Biology, 6(6): e1000815.
Hart, Anne C (2005), Behavior (July 3, 2006),
WormBook, Ed. The C. Elegans Research
Community, WormBook, doi/10.1895/wormbook.
1.87. 1.
Hucka, M., Finney, A., Sauro, H. M., Bolouri, H., Doyle,
J. C., Kitano, H., Forum, and the rest of the SBML,
Arkin, A. P., Bornstein, B. J., Bray, D., Cornish-
Bowden, A., Cuellar, A. A., Dronov, S., Gilles, E. D.,
Ginkel, M., Gor, V., Goryanin, I. I., Hedley, W. J.,
Hodgman, T. C., Hofmeyr, J.-H., Hunter, P. J., Juty,
N. S., Kasberger, J. L., Kremling, A., Kummer, U.,
Novère, N. Le, Loew, L. M., Lucio, D., Mendes, P.,
Minch, E., Mjolsness, E. D., Nakayama, Y., Nelson,
M. R., Nielsen, P. F., Sakurada, T., Schaff, J. C.,
Shapiro, B. E., Shimizu, T. S., Spence, H. D., Stelling,
J., Takahashi, K., Tomita, M., Wagner, J. and Wang, J.
(2003), The Systems Biology Markup Language
(SBML): A Medium for Representation and Exchange
of Biochemical Network Models. Bioinformatics,
19(4): 524–531.
Iersel, Martijn P. van, Villéger, Alice C., Czauderna,
Tobias, Boyd, Sarah E., Bergmann, Frank T., Luna,
Augustin, Demir, Emek, Sorokin, Anatoly, Dogrusoz,
Ugur, Matsuoka, Yukiko, Funahashi, Akira, Aladjem,
Mirit I., Mi, Huaiyu, Moodie, Stuart L., Kitano,
Hiroaki, Novère, Nicolas Le and Schreiber, Falk
(2012), Software Support for SBGN Maps: SBGN-ML
and LibSBGN. Bioinformatics, 28(15): 2016–2021.
Le Novère, Nicolas, Finney, Andrew, Hucka, Michael,
Bhalla, Upinder S, Campagne, Fabien, Collado-Vides,
Julio, Crampin, Edmund J, Halstead, Matt, Klipp,
Edda, Mendes, Pedro, Nielsen, Poul, Sauro, Herbert,
Input Encoding Proposal for Behavioral Experiments with a Virtual C. elegans Representation
119
Shapiro, Bruce, Snoep, Jacky L, Spence, Hugh D and
Wanner, Barry L (2005), Minimum Information
Requested in the Annotation of Biochemical Models
(MIRIAM). Nature Biotechnology, 23(12): 1509–
1515.
Le Novère, Nicolas, Hucka, Michael, Mi, Huaiyu,
Moodie, Stuart, Schreiber, Falk, Sorokin, Anatoly,
Demir, Emek, Wegner, Katja, Aladjem, Mirit I,
Wimalaratne, Sarala M, Bergman, Frank T, Gauges,
Ralph, Ghazal, Peter, Kawaji, Hideya, Li, Lu,
Matsuoka, Yukiko, Villéger, Alice, Boyd, Sarah E,
Calzone, Laurence, Courtot, Melanie, Dogrusoz, Ugur,
Freeman, Tom C, Funahashi, Akira, Ghosh, Samik,
Jouraku, Akiya, Kim, Sohyoung, Kolpakov, Fedor,
Luna, Augustin, Sahle, Sven, Schmidt, Esther,
Watterson, Steven, Wu, Guanming, Goryanin, Igor,
Kell, Douglas B, Sander, Chris, Sauro, Herbert,
Snoep, Jacky L, Kohn, Kurt and Kitano, Hiroaki
(2009), The Systems Biology Graphical Notation.
Nature Biotechnology, 27(8): 735–741.
Lloyd, Catherine M., Halstead, Matt D. B. and Nielsen,
Poul F. (2004), CellML: Its Future, Present and Past.
Progress in Biophysics and Molecular Biology, 85(2–
3): 433–450.
Mueller, Shane T. and Piper, Brian J. (2014), The
Psychology Experiment Building Language (PEBL)
and PEBL Test Battery. Journal of Neuroscience
Methods, 222: 250–259.
Neurobehavioral Systems (2015), Precise, Powerful
Stimulus Delivery. available at http://www.neurobs
.com/ [23 September 2015].
NuML Project (2015), NuML - Numerical Markup
Language. available at https://github.com/numl/numl
[23 September 2015].
OpenWorm Project (2015), OpenWorm Movement
Validation Project. available at https://github.
com/openworm/movement_validation [22 September
2015].
Schindelman, Gary, Fernandes, Jolene S., Bastiani, Carol
A., Yook, Karen and Sternberg, Paul W. (2011),
Worm Phenotype Ontology: Integrating Phenotype
Data within and beyond the C. Elegans Community.
BMC bioinformatics, 12: 32.
Si elegans Consortium (2015a), Behavioural Experiment
Input Definition XML Schema. available at
https://github.com/Si-elegans/behavioural_experiment
_definition/blob/master/behaviourDesc.xsd.
Si elegans Consortium (2015b), Behavioral Experiment
Input Encoding Example. available at https://github.
com/Si-elegans/behavioural_experiment_definition/
blob/master/Examples/example_nebica2015.xml.
Szigeti, Balazs, Gleeson, Padraig, Vella, Michael,
Khayrulin, Sergey, Palyanov, Andrey, Hokanson, Jim,
Currie, Michael, Cantarelli, Matteo, Idili, Giovanni
and Larson, Stephen (2014), OpenWorm: An Open-
Science Approach to Modelling Caenorhabditis
Elegans. Frontiers in Computational Neuroscience, 8.
available at http://www.frontiersin.org/Journal
/Abstract.aspx?s=237&name=computational_neurosci
ence&ART_DOI=10.3389/fncom.2014.00137.
Teeters, Jeffrey L., Harris, Kenneth D., Millman, K.
Jarrod, Olshausen, Bruno A. and Sommer, Friedrich T.
(2008), Data Sharing for Computational Neuroscience.
Neuroinformatics, 6(1): 47–55.
Waltemath, Dagmar, Adams, Richard, Beard, Daniel A.,
Bergmann, Frank T., Bhalla, Upinder S., Britten,
Randall, Chelliah, Vijayalakshmi, Cooling, Michael
T., Cooper, Jonathan, Crampin, Edmund J., Garny,
Alan, Hoops, Stefan, Hucka, Michael, Hunter, Peter,
Klipp, Edda, Laibe, Camille, Miller, Andrew K.,
Moraru, Ion, Nickerson, David, Nielsen, Poul,
Nikolski, Macha, Sahle, Sven, Sauro, Herbert M.,
Schmidt, Henning, Snoep, Jacky L., Tolle, Dominic,
Wolkenhauer, Olaf and Le Novère, Nicolas (2011),
Minimum Information About a Simulation Experiment
(MIASE). PLoS Comput Biol, 7(4): e1001122.
Waltemath, Dagmar, Adams, Richard, Bergmann, Frank
T., Hucka, Michael, Kolpakov, Fedor, Miller, Andrew
K., Moraru, Ion I., Nickerson, David, Sahle, Sven,
Snoep, Jacky L. and Novère, Nicolas Le (2011),
Reproducible Computational Biology Experiments
with SED-ML - The Simulation Experiment
Description Markup Language. BMC Systems
Biology, 5(1): 198.
Ward, Alex, Liu, Jie, Feng, Zhaoyang and Xu, X Z Shawn
(2008), Light-Sensitive Neurons and Channels
Mediate Phototaxis in C. Elegans. Nature
Neuroscience, 11(8): 916–922.
Wimalaratne, S. M., Halstead, M. D. B., Lloyd, C. M.,
Cooling, M. T., Crampin, E. J. and Nielsen, P. F.
(2009), A Method for Visualizing CellML Models.
Bioinformatics, 25(22): 3012–3019.
NeBICA 2015 - Symposium on Neuro-Bio-Inspired Computation and Architectures
120
