Sharing Knowledge in Daily Activity: Application in Bio-Imaging
Cong Cuong Pham
1
, Nada Matta
2
, Alexandre Durupt
1
, Benoit Eynard
1
, Marianne Allanic
1,4
,
Guillaume Ducellier
2
, Marc Joliot
3
and Philippe Boutinaud
4
1
Sorbonne University, University of Technology of Compiegne, Department of Mechanical Systems Engineering,
UMR CNRS 7337 Roberval, CS 60319, 60203 Compiegne Cedex, France
2
University of Technology of Troyes, 12 Rue Marie Curie, 10010 Troyes, France
3
GIN UMR 5296, CNRS, CEA, Bordeaux University, Case 71,146 rue Lo-Saignat, 33076 Bordeaux Cedex, France
4
Cadesis, 37 rue Adam Ledoux, 92400 Courbevoie, France
Keywords:
Knowledge Sharing, Ontology Product Life Cycle Management, Bio-Imaging.
Abstract:
Our approach uses the ontology to facilitate the data querying of users in the domain Bio-Imaging where
the data resources are heterogeneous and complex. The dependencies among data and the evolution of data
resources challenge users (especially for non-technician users) in querying the right data. Ontology can beused
to share the users’ understanding about data relationships to all community as well as to trace the database
evolution. As consequence, using ontology is a promising solution to facilitate the user’s query making process
and to enhance the query’s results.
1 INTRODUCTION
The evolution of data resources (new added data, new
generated relationships among data) challenges users
in data querying. In the context of distributed and het-
erogeneous data resources, since the new data com-
monly belong to an individual or group of individuals,
the others don’t understand the meaning of them as
well as the relationships between them and the exist-
ing data. The data querying becomes more and more
difficult.
This inconvenience can be overcome by using on-
tology as a method to assimilate the understanding
about data and their relationships to all users. In
this paper, we present an ontology-based query sys-
tem which facilitates the query making process of
scientists in the domain of Bio-Imaging where the
heterogeneity and complexity of data resources have
been handled by using Product Lifecycle Manage-
ment (PLM) solutions (Allanic et al., 2013). Our
query system added a semantic layer between the sci-
entists and PLM system, which helps each user to
query the database without helps of database techni-
cians.
The rest of paper is organized as follows: The sec-
tion 2 and 3 deal with some related works in knowl-
edge sharing using ontology and the data querying in
PLM system. Then, in section 4 and 5, we are going
to present our ontology-based approach and its appli-
cation. The section 6 is reserved for discussion and
conclusion.
2 DATA QUERYING IN
BIO-IMAGING
During quotidian activities, researchers in Bio-
Imaging domain manipulate heterogeneous data like
human information, brain images (2D, 3D), diag-
nostic information ... They produce then new im-
ages, statistics data, diseases description. Informa-
tion grows quickly and they need a support that han-
dles the data evolving more efficiently than the ex-
isting database. At Gin (Groupe dImagerie Neuro-
fonctionnelle UMR CNRS 5296) Lab (Bio-Imaging
lab) researchers use Product Life-cycle Management
(PLM) solutions. By definition, a PLM system con-
sists of tools that enable the management of the
whole product data and related information thought
all phases of product lifecycle (Eynard et al., 2004).
It has been proven as an efficient solution to tackle the
heterogeneity, complexity and the growth of data re-
sources to tackle the complexity, variety, heterogene-
242
Pham, C., Matta, N., Durupt, A., Eynard, B., Allanic, M., Ducellier, G., Joliot, M. and Boutinaud, P..
Sharing Knowledge in Daily Activity: Application in Bio-Imaging.
In Proceedings of the 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2015) - Volume 3: KMIS, pages 242-247
ISBN: 978-989-758-158-8
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
ity and growth of data resources.
In Bio-Imaging field, for a new study, a scientist
needs not only the original data but also the processed
data of others in order to enhance the study results. It
is important to help the scientists to query himself the
database. However, the traditional PLM systems are
not enough flexible, the data querying is only reserved
for database technicians.
3 KNOWLEDGE SHARING AND
PRODUCT LIFECYCLE
MANAGEMENT
3.1 Knowledge Sharing
Knowledge plays an important role in the long term
sustainability and success of organization. The need
for processes that facilitate the creation, sharing and
leveraging of individual and collective knowledge has
emerged recently for this reason. Knowledge sharing
(KS) has been introduced as one of the major activi-
ties of knowledge management and some definitions
of knowledge sharing can be found in the literature
(Small and Sage, 2006), (Hendriks, 1999). Some au-
thors (Sato et al., 2002), (Zhang et al., 2008) have
invested their efforts to construct platforms that en-
able knowledge sharing by using Information technol-
ogy ITs.(Sato et al., 2002) used XML Linking Lan-
guage (XLink) as a method of knowledge represen-
tation describing and proposed architecture for shar-
ing that knowledge among users. (Zhang et al., 2008)
tried to re-define knowledge resources in the network
by object-oriented thinking and proposed three-layer
knowledge sharing model. By using technologies on
Web 2.0, a knowledge-sharing system is built on the
Internet, allows the knowledge acquisition, sharing,
extension and retrieving.
Ontology is defined as an explicit formal specifi-
cation of a shared understanding. It has been used in
many knowledge sharing systems. For instance, (Yoo
and No, 2014) proposed a system based on ontol-
ogy expressing economics knowledge and Semantic
Web technologies (Domingue et al., 2011). This sys-
tem consists of five layers: registration, ontology, data
storage, reasoning and economic knowledge sharing.
Users can register economics knowledge pertaining
to a certain economics paper. They define then the
metadata and the relationships between notions dis-
cussed in the paper. According ontology model, the
system transforms this knowledge into semantic data
in a machine-understandable format. Two main func-
tions are basic search and knowledge navigation.
OntoShare (Davies et al., 2002) is another
ontology-based knowledge sharing. In his approach,
as users contribute information to the community, a
knowledge resource annotated with metadata is cre-
ated by using ontologies that have been predefined
using Resources Description Framework Schema
(RDFS) and populated using RDF.
Some authors have successfully applied ontology
as a method for knowledge sharing, but they did not
consider the complexity of relationships between data
and related information, in order to enhance the data
querying. Before going to propose our approach that
uses ontology to share knowledge in PLM, let us to
describe some main features of PLM.
3.2 Product Lifecycle Management
Product Life-cycle Management (PLM) systems
integrate constantly all the information produced
throughout all phases of a product’s life-cycle to ev-
eryone in an organization at every level (managerial,
technical) (Sudarsan et al., 2005). We can figure some
key advantages of PLM systems:
• An effective PLM system reduces enormous data
resources to coherentdata flows, avoids redundan-
cies.
• PLM permits the product structure management,
its evolution as well as the performed modifica-
tions tracking.
• PLM enables the collaboration through virtual,
distributed and extended enterprises.
• PLM tackles the heterogeneity, complexity of the
data resources as well as the confidentiality and
traceability issues.
However, along with these advantages, it also ex-
ists some issues:
• The lack of a standard between PLM systems
causes data integrity problems and limits the ac-
cess to and sharing of distributed product infor-
mation and knowledge .
• The increasing of need for product life-cycle
knowledge capitalization and reuse in order to re-
duce time and cost.
• PLM systems are not enough flexible and the data
querying requires a good understanding about
data model. This issue challenges non-technicians
users in data exploitation.
The challenges for end-users (scientists for ex-
ample) come from the low-level expression of data
model as well as the evolution of database. In this
Sharing Knowledge in Daily Activity: Application in Bio-Imaging
243
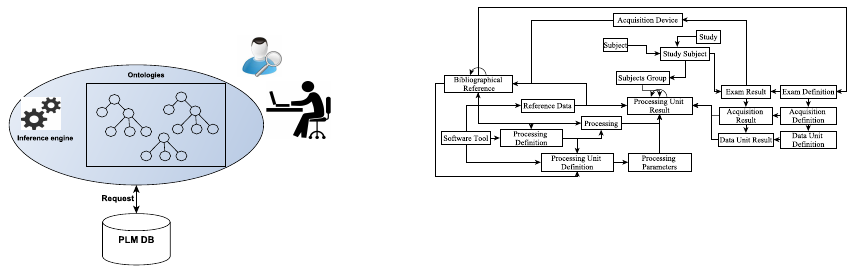
paper, our approach used ontology to share the un-
derstanding about database structure and the depen-
dencies among data to all types of users in order to
enhance the data querying.
4 ONTOLOGY-BASED
APPROACH IN BIO-IMAGING
4.1 Bio-Imaging Data Management in
PLM
The bio-imaging data have been handled in PLM
database Teamcenter 9.1. We have interviewed some
researchers at GIN lab to identify their needs and dif-
ficulties in manipulating with information system dur-
ing their quotidian activities. The results have shown
that most of scientists had difficulties in data querying
due to the lack of understanding about data model and
data relationships. They almost cannot accomplish
this task without helps of database technicians. Pro-
viding an efficient query interface for non-technicians
therefore becomes crucial.
Data querying becomes more and more complex
since each user introduces his own view when he/she
adds new data. Knowledge sharing techniques (on-
tology for example) we presented above help to de-
scribe the meaning of new data and relationships (an-
notations, descriptions) on the one hand to respect the
logic of a user and on the other hand to share knowl-
edge. We believe that our ontology-based approach
and inference engine will improve the data querying
of users at GIN (Figure 1).
Figure 1: Ontology-based knowledge sharing architecture
in PLM.
4.2 Ontology for Bio-Imaging
The main objective of using ontology is to facilitate
the data querying of Bio-Imaging scientists at GIN.
So, the concepts in ontology model need to refer
to concepts in the data model, and the lowest-level
concepts have to be linked to real data in the PLM
database.
4.2.1 BMI Data Model and Classification
Teamcenter
Beside the data model, Teamcenter 10 provides a tool
to classify data into categories. Some queries can be
executed by using this classification. Our ontology
has been built based on both data model and this clas-
sification.
Figure 2 presents the BMI-LM (Bio-Medical
Imaging - Lifecycle Management) data model used
in the PLM TeamCenter 9.1 (Allanic et al., 2013).
By adopting PLM solutions in the context of Bio-
Imaging, this PLM-oriented data model covered the
whole stages of a BMI study from specifications to
publications and enabled the flexibility in data man-
agement. It contains three types of objects:
• Definition objects (Exam Definition Data Unit
Definition, Acquisition Definition, Processing
Definition,) represent the definition that are used
to keep the traceability of data provenance during
the whole lifecycle of a study.
• Result objects: Acquisition Result, Data Unit Re-
sult, Exam Result, Processing Result.
• Result objects (Acquisition, Data Unit, Exam,
Processing) consist all acquired data during a par-
ticular study.
• Reference objects (Bibliographical, Reference
Data) concern data that can be defined and whose
can be represented device or system in the time.
Figure 2: BMI-LM DM implemented in Teamcenter 9.1.
Definition concepts have been created for the pur-
pose of data reuse. Some data have been acquired
under some conditions or by following a special pro-
tocol. These conditions and protocol are defined in
a definition concept to trace the provenance of data.
For example, all the Processing results computed by
using the same Acquisition device and the Processing
parameter can be attached to the same corresponding
Processing definition.
The classification (Figure 3) has been built based
on the data model. From that, BMI data have been
KMIS 2015 - 7th International Conference on Knowledge Management and Information Sharing
244
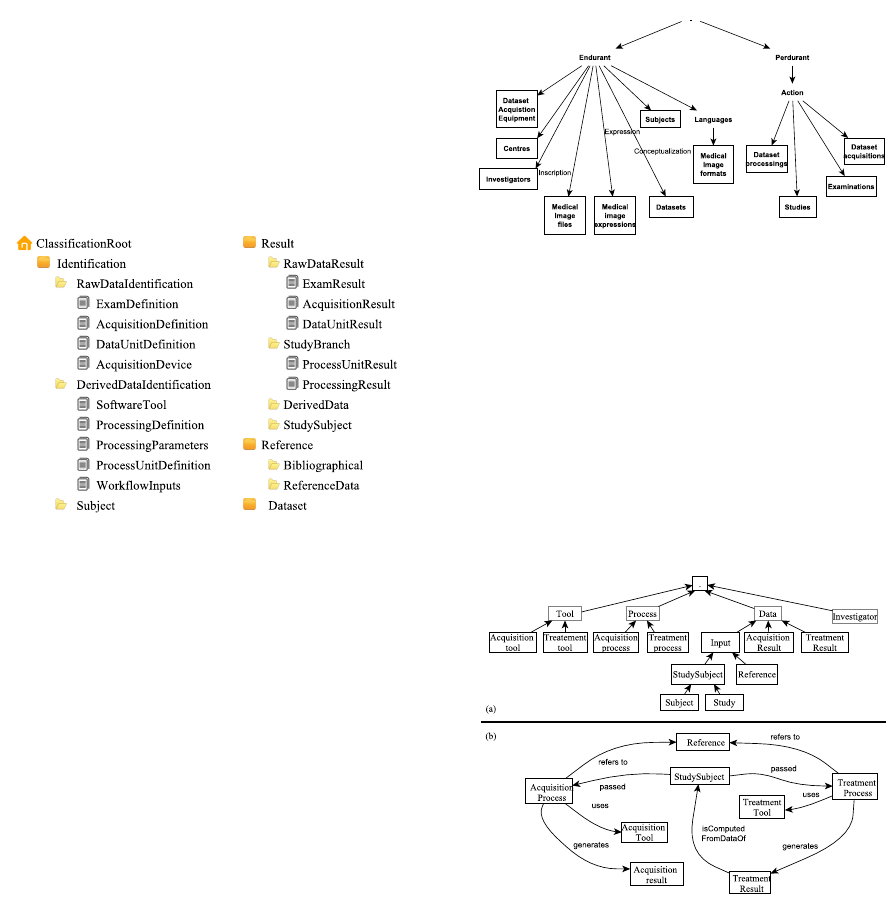
classified into branch, classes and sub-classes. The
classification allows a specific class to be added to a
generic item (object in the data model). At lowest-
level, data in PLM database have been organized in
tables based on the data model. The structure of clas-
sification, the classes’ attributes can be modified due
to the evolution of data resource. It provide the flex-
ibility capacity to users when they manipulate with
data (Allanic et al., 2013).
Figure 3: Classification in Teamcenter 9.1 corresponding
with the BMI-LM data model.
However, the nature of data in this classification
can be eventually repeated. It means some data can be
classified in two difference classes. This issue make
users confusing when they make a query. Further-
more, when the number of classes, sub-classes grows,
the relationships among these classes become com-
plex and can be not handled with out annotation.
To overcome this issue, we build an ontology,
which bases on both of data model and classification.
This ontology shows the logic of information in Bio-
Imaging and it provides an overview of all concepts
and relationships in the data model as well as in the
classification.
4.2.2 Ontological Model Construction
There is a lot of ontology defined in medicine,
for instance, in oncology, in neurology... (Gibaud
et al., 2011). But little works focus on Bio-Imaging
field. (Gibaud et al., 2014) defined concepts used in
Bio-Imaging like: Dataset, Processing, Investigators,
Medical Image files, Equipment, subjects, etc. (Fig-
ure 4)
Based on the data model BMI-LM and the clas-
sification in Teamcenter, we adapt this ontology to
the logic use of information in database. Infor-
mation belongs to three major categories: Tool,
Data and Process corresponding with acquisition de-
vice/software tool, acquisition/processing results, ac-
quisition/processing definition (Figure 5). This hi-
Figure 4: Ontology for Bio-Imaging (Eynard et al., 2004).
erarchy is more logic and understandable by non-
technicians users.
We define then relationships/restrictions among
concepts and sub-concepts like Uses, Generates, In,
Refers to, Passed For example, StudySubject have
passed Acquisition Process and this process gener-
ates some Acquisition Results. These relationships
are identified from the interview with scientists and
they reflect their work logic.
In the next section, we are going to present the
ontology-based query system as the application of our
approach.
Figure 5: Ontology at GIN: a) Ontology tree b) Ontology
graph.
5 ONTOLOGY-BASED QUERY
SYSTEM
Using ontology facilitates users in query making pro-
cess. The ontology tree and graph help users to under-
stand the relationships among concept and to directly
define query parameters. User can also chose a query
in the list of history queries to re-execute, modify or
complete it. Figure 6 illustrates the architecture of our
query-system.
Sharing Knowledge in Daily Activity: Application in Bio-Imaging
245
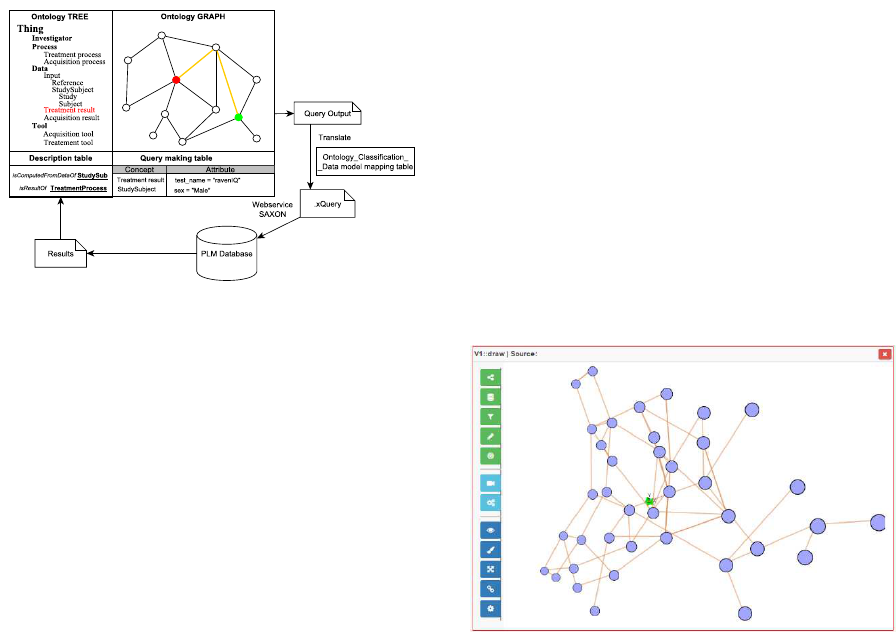
Basically, to query data in PLM database, user can
do as follows:
1. User identifies query concept and related query
concept in the ontology tree or graph.
2. In the description table, user sees all information
about current selected concept: Annotation, con-
cept usage, attributes and relationships (restric-
tions).
3. From the query concept, user navigates to all oth-
ers related concepts by following the predefined
restrictions. At each related concept, user can de-
fine the value for its attributes.
4. The step 3 is repeated until all related concepts
have been chose.
5. A mapping table between ontology concepts and
objects in data model will be used to automati-
cally generate a query which can be understand-
able and executable by the query engine.
6. The results will be displayed in the same query
interface, in a table or a graph.
We would to take here an example of query fre-
quently used by scientist at GIN lab:
Figure 6: Query making process using ontology.
Query all the results of RaventIQ test that was
performed on all male and right-handed subjects
who passed a T1 scan acquisition.
Here, RaventIQ is a test in Treatment Process
(Processing), while T1 scan is a test in Acquisition
Process. These tests have been performed on human
(StudySubject - A Subject in a predefined Study)
(Figure 2). If a researcher tries to use SQL to answer
this question, he has to know in which tables these
data saved and of course the relations among tables.
This is the SQL statement made by database techni-
cians:
SELECT sujet.codesujet, test.etiquette,
passation.valeur, lateralite.main_ecrit,
fichier.nomfichier, sujet.sexe
FROM sujet, passation, lateralite, test,
examen, acquisition, fichier
WHERE sujet.id = passation.sujet
AND sujet.id = examen.sujet
AND sujet.id = lateralite.sujet
AND examen.id = acquisition.examen
AND acquisition.fichier = fichier.id
AND passation.test = test.idtest
AND passation.valeur NOT LIKE ’’
AND test.etiquette = "ravenQi"
AND lateralite.main_ecrit = ’D’
AND fichier.nomfichier LIKE ’%_t1_%’
AND sujet.sexe = ’H’
ORDER BY sujet.codesujet
However, the query construction process can be
simplified by using ontology. Each scientist knows
that he can find the results of RaventIQ test in Treat-
ment Result, so he chooses concept Treatment Result
in Ontology Tree (or graph) as the query concept.
From Description Table, he know that this concept is
related to StudySubject concept by property: isCom-
putedFromDataOf. He chooses this concept and sets
the value Male for subject’s sex attribute. Here, he
will see that a StudySubject has passed some Acquisi-
tion processes which generate Acquisition Results. He
continues choosing related query concepts and sets
the value for its attributes (right-handed and T1).
We use TCXquery, a tool that considers PLM data
as XML documents, as the Query Processor. So when
user finishes, a query will be automatically generated
in .xquery format by using a table that maps a restric-
tion between two ontology concepts to a predefined
route between two objects in data model. With this
mapping table, users don’t have to know previously
the data model. At the end, .xquery query will be sent
to and executed at server by using a web service (Fig-
ure 6).
Figure 7 illustrates our query interface where the
results obtained are represented as graph in the same
interface in order to facilitate the sharing of data and
information among users for further purpose.
Figure 7: Ontology-based query system in Bio-Imaging.
KMIS 2015 - 7th International Conference on Knowledge Management and Information Sharing
246
6 CONCLUSION
In this paper, we presented an ontology-based query
system that facilitates the query making of users in
Bio-Imaging domain. The main component of our ap-
proach is the ontological model and the table mapping
between this ontology and the data model in PLM
system. Another important factor is the relationships
definition among concepts of ontology. It reflects on
the one hand the logic work of users and on the other
hand, it determines the accuracy of our approach.
As future work we will focus on the test of
proposed query interface with various queries sets
(in BioImaging domain) and engineering design (in
PLM). The ontology tree and ontology graph must be
also developped to cover all concepts in BioImaging
domain. Ontology will be implemented in semantinc
web language (RDF, SPARQL) in order to more use
inference engine for information search.
ACKNOWLEDGEMENTS
The work presented in the paper is conducted
within the ANR (Agence Nationale de la Recherche)
founded project BIOMIST (noANR-13-CORD-0007)
for the matic axis no2 of the Contint 2013 Call for
Proposal: from content to knowledge and big data.
REFERENCES
Allanic, M., Durupt, A., Joliot, M., Eynard, B., and Boutin-
aud, P. (2013). Application of PLM for Bio-Medical
Imaging in Neuroscience. In Bernard, A., Rivest, L.,
and Dutta, D., editors, Product Lifecycle Management
for Society, number 409 in IFIP Advances in Informa-
tion and Communication Technology, pages 520–529.
Springer Berlin Heidelberg.
Davies, J., Duke, A., and Stonkus, A. (2002). OntoShare:
Using Ontologies for Knowledge Sharing. In Seman-
tic Web Workshop, volume 72.
Eynard, B., Gallet, T., Nowak, P., and Roucoules, L. (2004).
UML based specifications of PDM product structure
and workflow. Computers in Industry, 55(3):301–316.
Gibaud, B., Forestier, G., Benoit-Cattin, H., Cervenan-
sky, F., Clarysse, P., Friboulet, D., Gaignard, A.,
Hugonnard, P., Lartizien, C., Liebgott, H., Montag-
nat, J., Tabary, J., and Glatard, T. (2014). OntoVIP:
An ontology for the annotation of object models used
for medical image simulation. Journal of Biomedical
Informatics, 52:279–292.
Gibaud, B., Kassel, G., Dojat, M., Batrancourt, B., Michel,
F., Gaignard, A., and Montagnat, J. (2011). Neu-
roLOG: sharing neuroimaging data using an ontology-
based federated approach. AMIA ... Annual Sympo-
sium proceedings / AMIA Symposium. AMIA Sympo-
sium, 2011:472–480.
Hendriks, P. (1999). Why share knowledge? The influ-
ence of ICT on the motivation for knowledge sharing.
Knowledge and Process Management, 6(2):91–100.
Sato, H., Otomo, K., and Masuo, T. (2002). A knowl-
edge sharing system using XML Linking Language
and peer-to-peer technology. pages 26–27.
Small, C. T. and Sage, A. P. (2006). Knowledge manage-
ment and knowledge sharing: A review. Information,
Knowledge, Systems Management, 5(3):153–169.
Sudarsan, R., Fenves, S. J., Sriram, R. D., and Wang, F.
(2005). A product information modeling framework
for product lifecycle management. Computer-Aided
Design, 37(13):1399–1411.
Zhang, J., Liu, Y., and Xiao, Y. (2008). Internet Knowledge-
Sharing System Based on Object-Oriented. In Second
International Symposium on Intelligent Information
Technology Application, 2008. IITA ’08, volume 1,
pages 239–243.
Sharing Knowledge in Daily Activity: Application in Bio-Imaging
247
