
Teaching & Learning System for Diagnostic Imaging
Phase I: X-Ray Image Analysis & Retrieval
M. S. Shahriar Faruque, Shourav Banik, M. Kazi Mohammed, Mahady Hasan and M. Ashraful Amin
Computer Vision and Cybernetics Group, Department of CSE, Independent University,
Bangladesh, Dhaka-1229, Bangladesh
Keywords: Diagnostic Imaging, X-Ray, Medical Image Annotation, Semi-Auto Segmentation, Information Retrieval,
Image Retrieval.
Abstract: This paper presents a framework for building diagnostic imaging teaching and learning facility for entry level
medical students of Bangladesh. Initially we demonstrate an X-Ray image analysis and retrieval system that
will work as one of the main component in this system. This web based system has three modes. First is the
annotation mode where an expert radiologist manually performs annotation of raw x-ray images. To aid the
annotation process proposed model proposes a manual and a semi-auto segmentation tool in identifying the
region of interests (ROI) in the X-Ray images. Image Retrieval in Medical Applications (IRMA) structure
has been used for the annotating the ROIs. In the learning mode, students can retrieve images from the
database created by expert radiologists. We proposed information retrieval techniques to find x-ray images of
interest. We have used text based and content based search methods which is based on term frequency–inverse
document frequency (tf-idf), and Gabor filter respectively.
1 INTRODUCTION
Bangladesh is a densely populated country with a
population of 156.06 million. A country with this
much population always needs a strong medical
sector and this country is no exception. In order to
reduce mortality rate and provide a good health
support government is constantly trying to expand
and improve the medical sector. There are currently
592 government and 2983 private registered hospitals
in this country according to the recent health bulletin
(Directorate General of Health Services, 2014).
Medical experts are always of need. To be an expert
in any medical field one needs rigorous amount of
practice and guidance. Medical colleges do provide
such training programs and guidance. Along with
these facilities, internet has become very popular in
this regard, as one can find a tremendous amount of
information on it. Then again such resources are
scattered and needs validation. So an authentic self-
sufficient system is needed to address these needs.
Medical imaging is one such sector which lacks a
proper teaching and learning system. Thus a system
which will enable the trainees to learn and assess their
knowledge in medical imaging is of need. Some
systems do exist but they are expensive; thus hard for
most of the students to acquire. A teaching and
learning system such as this has three distinguished
part. They are: teaching and learning models, medical
image annotation, and image retrieval.
In order to build our own teaching and learning
system for diagnostic imaging we are proposing a
model, composed of three different interfaces: expert
mode, learning mode, and exam mode. Expert mode
is where expert radiologists will annotate different
diagnostic images. This will in turn, build our
database of complete annotated diagnostic images.
Along with basic geometric shapes, a semi-automatic
detection tool has been introduced here to select
various regions of interests (ROI) in images. To
annotate the ROI a well-organized classification
structure is required. For this purpose, we decided to
follow IRMA coding system. IRMA coding system
specializes in classification of medical images.
Secondly in learning mode trainees can view
annotated images from the dataset and search for
specific type of radiographs. For searching we have
integrated two methods: text based (TBIR) and
content based (CBIR) image retrieval. Lastly we have
built an exam mode so that students can assess
430
Faruque M., Banik S., Mohammed M., Hasan M. and Amin M..
Teaching & Learning System for Diagnostic Imaging - Phase I: X-Ray Image Analysis & Retrieval.
DOI: 10.5220/0005479604300435
In Proceedings of the 7th International Conference on Computer Supported Education (CSEDU-2015), pages 430-435
ISBN: 978-989-758-107-6
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

themselves by trying to correctly annotate body parts
in diagnostic images.
Much work has already been done on this field. In
2003, IRMA structure was proposed which uses a
mono-hierarchical multi-axial classification code
which preserves the advantages of SNOMED
DICOM and also provide advantage in content based
image retrieval (Lehmann, Schubert, Keysers,
Kohnen & Wein, 2003). To select a region of interest
geometric tools was proposed by Schneider and
Eberly (2003). Barrett and Mortensen introduced a
semi-auto segmentation tool which was capable of
segmenting the region of interest using very little time
and effort (1997).
For building medical image retrieval system a
hierarchal similarity learning method using neural
networks and support vector machines was proposed
by El-Naqa, Yang, Galatsanos, Nishikawa & Wernick
(2004). An automatic indexing and retrieval method,
based on medical concept from the Unified Medical
Language System (UMLS) was recommended for an
image retrieval system. To learn semantics from
images, a vector machine is used as a structured
learning framework. In order to parse a XML
database where tags can have multiple meaning a
novel XML TF*IDF ranking strategy was proposed
(Bao, Lu, Ling & Chen, 2010). Avni, Greenspan,
Konen, Sharon and Goldberger represented image
contents by local patch using Bag-of-Words
model (BoW model). Nonlinear kernel-based Support
Vector Machine (SVM) was used to classify images.
The system was able to successfully discriminate
between healthy and pathological chest radiographs
(Avni, Greenspan, Konen, Sharon & Goldberger,
2011). To improve retrieval performance using
adaptive wavelet, a regression function is used which
estimates the best wavelet filter. For every possible
separable or non-separable wavelet filter,
image characterization is computed almost instantly
using an algorithm proposed by Quellec, Lamard,
Cazuguel, Cochener and Roux (2012). In order to
rank the search results of a query using text based
search, content based image search was proposed
(Cai, Zha, Wang, Zhang & Tian, 2014).
2 DATA
To examine the efficiency and usability of our system
we acquired radiographs from different hospitals in
Bangladesh. A total of 4324 DICOM images were
collected. The DICOM images were downscaled to
JPEG images preserving 80% or above image quality.
Each image had a fixed width of 1600 pixels and the
height was proportioned accordingly to maintain the
original aspect ratio. All of the images were then
sorted manually into head-neck, body, lower limb,
upper limb. Radiographs without any human body
parts were discarded into true negative category. The
sorting was done for the convenience of working.
This sorting does not influence the search methods.
The manual sorting process revealed some
interesting information about the dataset. It became
clear that not all radiographs were perfectly taken.
Some radiographs had a couple of problems including
blurriness and region out of focus.
3 PROPOSED MODEL
In order to create the database for our proposed
system, we have created a user interface where
experts can load any jpeg images. This software
includes tools for selecting specific region of interest
in the image and annotate them accordingly. The
information given in this front end would then be
stored in an xml file. As xml tags are generic these
can be used in almost any system. We have built the
annotating part of images around the IRMA coding
structure. After annotating an image a user can see
that image from the front end with annotations. A
main focus of this system is searching for images in
our database. In that regard two types search tool have
been integrated. Text based searching uses the tags
created for annotations. It implements a modified
version of tf-idf method. Content based search uses
Gabor filter to search any image from a given image.
Also we have added an exam mode for comparing
annotations automatically. This is based on each
annotated segment of an image. Further elaborations
on these are given below. Figure 1 illustrates the front
end of this system.
3.1 Expert Mode
Expert mode consists of tools to help the annotation
of radiographs. By using this interface expert
radiologists are able to annotate selected radiographs.
Expert Mode uses an elegant annotation system and a
set of selection tools to efficiently annotate the region
of interest. After the annotation is finished the data is
stored as an xml structure. A help button is included
to guide a new user throughout annotation the
process. Also a magnification tool is included so the
expert can observe the fine detail in the radiographs
and make annotations accordingly. Lastly the expert
can backtrack and view previous annotations by
pressing the show annotation button.
Teaching&LearningSystemforDiagnosticImaging-PhaseI:X-RayImageAnalysis&Retrieval
431

Figure 1: Front End.
3.1.1 Manual Selection Tool
For selecting different segment of the image, we built
manual and semi-auto selection tools. The manual
tool comprises of basic polygon, ellipse, point and
line. In spite of having a semi auto detection tool we
decided to keep these basic shapes as an alternate
way. If for some reason semi auto selection tool fails
to select a certain part of image then the expert can
manually select his desired region. This type of cases
may arise because of poor image quality. Using the
polygon almost any kind of shape can be selected
within the image. Ellipse is a fast way of selecting a
gross area. Lines can be used to identify decay of
bones. Figure 2 shows us an image which has been
Figure 2: Manual Selection Tool.
segmented by using the manual selection tool. This
structure is created using matlab’s built in functions.
The x and y coordinates of the structure are stored in
the xml file along with their respective annotations.
3.1.2 Semi Auto Selection Tool
Though in some context manual segmentation is
useful it has its flaws. One of which is imprecision.
Background and many unwanted pixels are included
in the manual segmentation. To avoid this after
manual segmentation a NN-clustering approach can
be used in some cases. But acquiring optimal
segmentation is still a problem because clustering
requires a predefined knowledge about neighbours.
Also it is time consuming and impractical when
applied to extensive spatial and temporal sequence of
images. To overcome these problems a semi-auto
segmentation tool livewire is included in the expert
mode. Livewire uses local cost function to determine
pixel to pixel travel cost for each pixel of the image
which is a weighted sum of the laplacian zero-
crossing, f
Z
, gradient magnitude, f
G
, and gradient
direction, f
D
. The local cost function,
l(p,q)= ω
G
* f
G
(q)+ ω
Z
* f
Z
(q)+ ω
D
* f
D
(p,q) (1)
Here l (p,q) means local cost for the directed edge
from pixel p to a neighbouring pixel q. The value of
weight ω
G
, ω
Z
and ω
D
is set to 0.43, 0.43 and 0.14
respectively. Then directed graph search is used to
find the shortest (minimal cost) path between the start
point and the seed point (Barrett & Mortensen, 1997).
The image maintaining aspect ratio is downscaled to
increase the speed of the segmentation tool. The
downscale ratio is then multiplied with the extracted
points to map the extracted points to the original copy
of the image. Figure 3 gives an example of this tool.
Figure 3: Semi Auto Selection Tool.
CSEDU2015-7thInternationalConferenceonComputerSupportedEducation
432

3.1.3 Annotation System
In order to systematically organize our annotations
we decided to follow the IRMA coding system. It is a
mono-hierarchical multi axial classification code
designed for medical images. IRMA structure was
used due to its rising popularity and efficiency. There
are four axes with three to four positions. Each
position is denoted by either digits ranging from 0 to
9 or alphabets ranging from a-z. The digit 0 represents
“unspecified” and determines the end of a path along
an axis. The four parts are: T (technical): image
modality, D (directional): body orientation, A
(anatomical): body region examined, B (biological):
biological system examined (Lehmann, Schubert,
Keysers, Kohnen & Wein, 2003). Besides this, we
also kept a field for the expert to write further
comments for a segment. After tagging the image, all
of the information is saved in an xml file. Additional
information like name, dimensions, expert comments
are also saved here. A partial xml file can be seen in
figure 4.
3.2 Learning Mode
The main purpose of creating this annotated image
database is to make the learning process easier. So
information retrieval is an integral part of this system.
From our learning window users can load any
annotated image. This will show them the image with
proper annotations. In this window users have the
option for initiating a text or context based search.
<root>
<name_of_file idx="1" type="char"
size=""></name_of_file>
<width idx="1" type="double" size="1 1"></width>
<height idx="1" type="double" size="1 1"></height>
<technique idx="1" type="struct" size="1 1">
<field1 idx="1" type="char" size=""></field1>
<field2 idx="1" type="char" size=""></field2>
<field3 idx="1" type="char" size=""></field3>
<field4 idx="1" type="char" size=""></field4>
</technique>
<wire idx="1" type="struct" size="">
<pos idx="1" type="double" size=""></pos>
<posx idx="1" type="double" size=""></posx>
<posy idx="1" type="double" size=""></posy>
<anatomy idx="1" type="struct" size="1 1">
<field1 idx="1" type="char" size=""></field1>
<field2 idx="1" type="char" size=""></field2>
<field3 idx="1" type="char" size=""></field3>
</anatomy>
</wire>
</root>
Figure 4: XML File.
3.2.1 Text based Search
Our text based search model is based on IRMA tags
and the comments given by experts. As all the tags of
an image are stored in an xml document, we
implemented a modified version of term frequency-
inverse document frequency (tf-idf) method
(Silberschatz, Korth & Sudarshan, 2006). This was
also used by Barrios, Diaz-Espinoza & Bustos (2009).
For our system, we did not use term frequency as it
failed to produce relevant result. Instead we
calculated the percentage of a segmented part in an
image. This was done by calculating total amount of
pixels of a segmented part, then this was divided by
the total number of pixels of the image. This acquired
value is used as our modified term frequency (tf’).
tf' = (p (d,t)) / (n (d)) (2)
Here, let p (d, t) be the total number of pixels for
tag t in xml document d and n (d) be the total number
of pixels in xml document d. IDF was achieved by
using the following formula
idf = log ( n(d) / n(d,t) )
(3)
Here, let n (d) be the total amount of xml
document in our database and n (d, t) be the amount
of documents where the term t is present. When a user
searches for tag using text based search appropriate
tf’ and idf values are generated and multiplied to get
the tf-idf value of documents. Then the images are
sorted according to this value and shown in a new
search result window as depicted in Figure 6.
Figure 5: Search Results.
Teaching&LearningSystemforDiagnosticImaging-PhaseI:X-RayImageAnalysis&Retrieval
433
3.2.2 Content based Search
In learning mode user can find similar images just by
selecting a specific image from the text-based search
result. Also the user can search similar images by
providing a reference images. Colour based feature
extraction systems is widely used in content-based
image retrieval system. But most medical images are
grey scale. So in most of the cases colour features will
not be useful. Shape-based feature extraction is most
preferable but it lacks granularity. So texture based
feature extractor Gabor wavelet is used as they can
potentially reflect the fine details contained within an
image structure (Akgül et al., 2010).
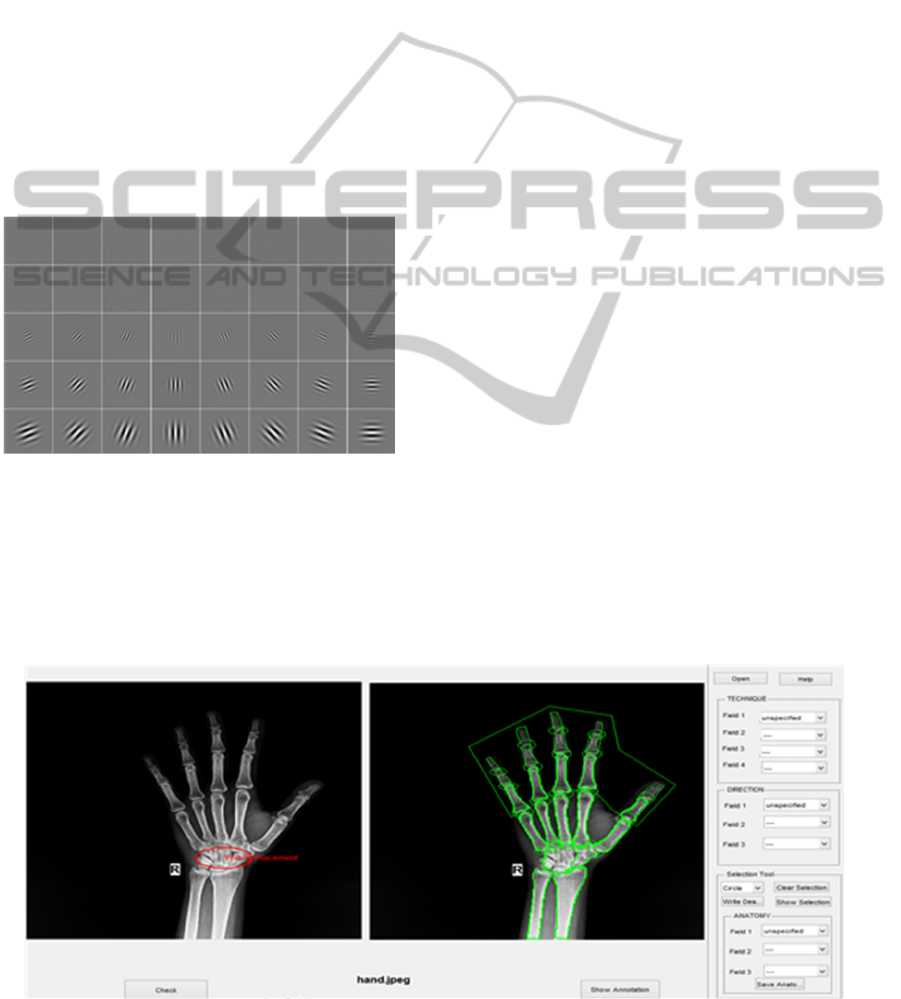
Gabor wavelet is used to generate feature vector
of 10000X1 dimension for each image which is
entered into the database. 40 filter of 5 different scales
and eight different direction is used to extract the
features. The filters are shown below in Figure 5.
Figure 6: Gabor Filters (left: real parts, right: magnitude).
Dimension of each filter is set to 39X39. The
feature vectors are stored in the database for
similarity measuring. When an image is provided its
feature vector of 100000X1 generated using the
Gabor wavelet. The feature vector is than compared
using Euclidian distance with the feature vectors in
the database. The result is than sorted and the top 20
results are then shown in the search window as
depicted in previously mentioned Figure 4.
3.3 Exam Mode
The exam mode is created so that the users can test
the accuracy of their annotation. Previous annotation
done by an expert radiologist is compared to the
annotation done by the user to give the user an
overview. This gives the user a chance to hone their
skill. The exam mode contains the same annotation
system and segmentation tools as the expert mode.
After annotating the image the user have to press the
check button. The result is shown in the right side of
the same window which shows if the user’s
annotation is wrong or correct. One of four possible
outcome is displayed for each selected region of
interest. The four outcomes are as follows.
Selected annotation exists for the image but the
enclosed region doesn’t match with the
respective region enclosed by the expert.
Selected annotation does not exist for the image
but the enclosed region matches with a region
enclosed by the expert.
Selected annotation does not exist for the image
and the enclosed region doesn’t match with any
region enclosed by the expert.
Selected annotation exists for the image and the
enclosed region matches with the respective
region enclosed by the expert.
Figure 7 shows the different outcomes for
selecting different regions of a selected radiograph.
The wrong annotations are shown in red while right
ones are shown in green. For matching regions when
showing one of four outcomes, a centroid is
calculated for a region enclosed by the user. A match
occurs when a centroid is inside a region annotated by
the expert.
Figure 7: Exam Mode.
CSEDU2015-7thInternationalConferenceonComputerSupportedEducation
434

4 CONCLUSIONS AND
FUTURE WORK
X-ray is a fundamental part of medical imaging. Any
trainee trying to become a radiologist will benefit
greatly from an X-ray annotation learning tool. Our
system which focuses solely on teaching and learning
radiograph can help such trainees greatly. Also
annotated radiographs from the experts can be further
used for research. This research work has the scope
for future work. We aim to introduce a completely
automatic annotation tool which will intelligently
annotate different body parts using the IRMA coding
structure. Also newer and better structures are being
introduced which can boost the accuracy of
annotations. Image retrieval system can be further
improved by adding a shape based search method.
REFERENCES
Akgül, C., Rubin, D., Napel, S., Beaulieu, C., Greenspan,
H. and Acar, B. (2010). Content-Based Image Retrieval
in Radiology: Current Status and Future Directions. J
Digit Imaging, 24(2), pp.208-222.
Avni, U., Greenspan, H., Konen, E., Sharon, M. and
Goldberger, J. (2011). X-ray Categorization and
Retrieval on the Organ and Pathology Level, Using
Patch-Based Visual Words. IEEE Transactions on
Medical Imaging, 30(3), pp.733-746.
Bao, Z., Lu, J., Ling, T. and Chen, B. (2010). Towards an
Effective XML Keyword Search. IEEE Transactions
on Knowledge and Data Engineering, 22(8), pp.1077-
1092.
Barrett, W. and Mortensen, E. (1997). Interactive live-wire
boundary extraction. Medical Image Analysis, 1(4),
pp.331-341.
Barrios, J., Diaz-Espinoza, D. and Bustos, B. (2009). Text-
Based and Content-Based Image Retrieval on Flickr:
DEMO. 2009 Second International Workshop on
Similarity Search and Applications.
Cai, J., Zha, Z., Wang, M., Zhang, S. and Tian, Q. (2014).
An Attribute-assisted Reranking Model for Web Image
Search. IEEE Trans. on Image Process., pp.1-1.
Directorate General of Health Services, (2014). Health
Bulletin 2014.
El-Naqa, I., Yang, Y., Galatsanos, N., Nishikawa, R. and
Wernick, M. (2004). A Similarity Learning Approach
to Content-Based Image Retrieval: Application to
Digital Mammography. IEEE Transactions on Medical
Imaging, 23(10), pp.1233-1244.
Lehmann, T., Schubert, H., Keysers, D., Kohnen, M. and
Wein, B. (2003). The IRMA code for unique
classification of medical images. Medical Imaging
2003: PACS and Integrated Medical Information
Systems: Design and Evaluation.
Quellec, G., Lamard, M., Cazuguel, G., Cochener, B. and
Roux, C. (2012). Fast Wavelet-Based Image
Characterization for Highly Adaptive Image Retrieval.
Schneider, P. and Eberly, D. (2003). Geometric Tools for
Computer Graphics. Amsterdam: Boston.
Silberschatz, A., Korth, H. and Sudarshan, S. (2006).
Database System concepts. 5th ed. Boston: McGraw-
Hill Higher Education.
Teaching&LearningSystemforDiagnosticImaging-PhaseI:X-RayImageAnalysis&Retrieval
435
