
Robust Image Analysis of BeadChip Microarrays
Jan Kalina
1
and Anna Schlenker
2,3
1
Dept. of Medical Informatics and Biostatistics, Institute of Computer Science,
Academy of Sciences of the Czech Republic, Prague, Czech Republic
2
Institute of Hygiene and Epidemiology, First Faculty of Medicine, Charles University in Prague, Prague, Czech Republic
3
Dept. of Biomedical Informatics, Faculty of Biomedical Engineering,
Czech Technical University in Prague, Kladno, Czech Republic
Keywords:
Microarray, Robust Image Analysis, Noise, Outlying Measurements, Background Effect.
Abstract:
Microarray images in molecular genetics are heavily contaminated by noise and outlying measurements. This
paper is devoted to analysis of Illumina BeadChip microarray images, primarily to their low-level preprocess-
ing. We point out that standard image analysis procedures, which are implemented in the beadarray package
of BioConductor software, are highly sensitive to contamination by severe noise and outliers. Therefore, the
habitually used methodology does not discover many of the outliers. We illustrate this on real data and show
that the standard background correction method may actually amplify the noise in the image. A robust image
analysis tailor-made for this type of microarray images is highly desirable. We explain principles and show
preliminary results of our robust alternative to the standard approach, which aims to be robust to noise and
outliers in each its step.
1 BEADCHIP MICROARRAYS
Microarrays represent a commonly used technology
for measuring gene expressions. Microarray studies
are typically designed to find differently expressed
genes in two or more groups of samples (e.g. patients
with a different form of a disease) or to perform (un-
supervised) clustering or (supervised) classification
analysis into given groups (Fraser et al., 2010).
Illumina BeadChip microarrays are claimed to be
the currently most popular technology for measuring
gene expressions (Rueda, 2014). A sample is placed
in a microwell array containing about a million of sil-
ica beads corresponding to different gene transcripts
(Kuhn et al., 2004). The surface of the chip is scanned
to obtain a gray-scale image with a high fluorescence
intensity corresponding to highly expressed genes.
We focus on the low-level preprocessing of Bead-
Chip images, which is a part of the BeadChip image
analysis with the aim to estimate expression valuesfor
each bead from the raw scanned image, i.e. after filter-
ing out the effect of the background. Various sources
call the low-level preprocessing by different names:
• Feature-intensity extraction (Smith et al., 2010)
• Low-level analysis (Dunning et al., 2008)
• Data reduction (Fraser et al., 2010)
• Quantification (Rueda, 2014)
The habitual approach to the image analysis of
BeadChip microarrays is based on the algorithm of
(Dunning et al., 2007), which does not contain a de-
scription of details of the methodology and practition-
ers have to rely on default values of its parameters.
The approach is implemented in the package beadar-
ray, which in its current version 1.16 can be con-
sidered a standard software for the preprocessing of
BeadChip microarrays, before performing a more ad-
vanced analysis with the standard lumi package. Both
packages are part of the open-source software Bio-
conductor (Rueda, 2014).
The basic intensity is observed everywhere in the
image and the target intensity in a particular bead cor-
responds to a gene expression. A raw BeadChip im-
age is a 16-bit gray-scale image, which contains one
of 65536 values. Performing the complex process of
outlier or noise detection is computationally intensive
and the difficult task of information extraction from
the massive 2D images with tens of millions of pixels
makes the field of microarrays image analysis to be
an important hot topic in current bioinformatics (Car-
dona and Tomancak, 2012).
Observed gray intensities of BeadChip microarray
images are heavily contaminated both in the beads as
89
Kalina J. and Schlenker A..
Robust Image Analysis of BeadChip Microarrays.
DOI: 10.5220/0005246900890094
In Proceedings of the International Conference on Bioimaging (BIOIMAGING-2015), pages 89-94
ISBN: 978-989-758-072-7
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

well as in the background. We have observed a con-
tamination by additive as well as impulsive (severely
outlying) noise in isolated pixels or in smaller or
larger regions in a variety of applications due to var-
ious reasons. To give only a few of them, these in-
clude measurement errors, unreliable sample prepa-
ration, or wrong identification of gene probes (Fraser
et al., 2010).
Only marginal attention has been paid to the low-
level preprocessing of BeadChip images. Specific
problems of BeadChips do not allow to simply use
sophisticated tools for image analysis and quality as-
sessment, which are available for Affymetrix microar-
rays (Kuhn et al., 2004). Instead, we hold the opinion
that a robust alternative tailor-made for BeadChip im-
ages would be highly desirable and we bring argu-
ments in favor of such opinion in this paper.
This paper has the following structure. Section 2
explains that procedures habitually applied on Bead-
Chip microarray images are highly vulnerable with
respect to noise. Section 3 proposes a computational
improvement for the foreground estimation. Cur-
rently, we work on robustifying standard image anal-
ysis approaches for BeadChip microarrays and our
ideas of robust image analysis for this task are sum-
marized in Section 4. Some results of the analysis of
real data are shown Section 5, revealing the sensitivity
of standard image analysis procedures with respect to
noise and outliers in the images. Finally, Section 6
concludes the paper.
2 PROBLEMS OF STANDARD
LOW-LEVEL PREPROCESSING
Standard analysis of BeadChip microarry images is
sensitive to random or systematic errors in the raw
data and to artifacts of different sizes and shapes. Its
methods havebeen derivedprimarily with the require-
ment on a high speed (Kuhn et al., 2004). This section
describes all steps of the standard image analysis al-
gorithm and our critical stance will be illustrated on
examples in Section 5. Thus, we bring new ideas and
findings beyond those of (Smith et al., 2010), who
simply recommended to ignore problematic beads in
the images.
Let us now critically describe all the steps of the
standard image analysis performed on each BeadChip
microarray image. Commonly, the same sample is ap-
plied on two neighboring strips (Shi et al., 2009). The
following are the steps implemented in the beadarray
package.
• Estimating the Local Background Effect by the
5-th smallest value in a square neighborhood of
each bead is heavily influenced by local noise.
• Estimating the Foreground is performed by
three consecutively applied linear filters.
1. Image Sharpening can yield negative intensi-
ties. Besides, it yields nonsense (also negative)
values for pixels at the boundary of the image
where there are no beads.
2. Averaging (mean filter) over a square of size
3× 3 pixels around each given bead propagates
the effect of outliers to their neighbors.
3. Another averaging of four neighboring pixels
of a particular bead.
• Background Correction defined as the differ-
ence between foreground and background inten-
sities. It may also yield negative values, which is
the consequence of the sharpening.
• Data Normalization has been largely discussed
and positively evaluated (Shi et al., 2009), but it
leads to hiding some outliers.
• Outlier Deletion is performed after mixing the
data from both strips, which prevents some out-
liers from being detected.
Unfortunately, these methods in the beadarray
package with default values of their parameters are
strongly influenced by noise and random or system-
atic errors in the measurements. They transfer the ef-
fect of noise from noisy pixels also to neighboring
pixels (and neighboring genes), which can be am-
plified from one non-robust step to later ones. In
this way, noise is introduced artificially to such genes
which are not affected by noise in the raw image. The
results of the whole process are also too much influ-
enced by the sharpening. Besides, there is not even an
attempt to correct for spatial artifacts.
Attention has been paid to the choice of a suitable
background correction method (Smith et al., 2010),
which has only a small influence on the result, but
no doubts have been cast upon other steps of the low-
level preprocessing. Nevertheless, they all can be eas-
ily shown to have a zero breakdown point, which is
a statistical measure of sensitivity against outliers in
the data (Davies and Gather, 2005).
The local approach to estimation of both back-
ground and foreground does not exploit information
about global trends across the whole strip. Besides,
the initial steps of the standard image analysis of mi-
croarrays are strongly influenced by local noise in the
neighborhood of particular microbeads and the result-
ing biased values are passed on to the next estima-
tion methods and transformations. Outliers become
masked among the data and their consequent detec-
tion and deletion is much more complicated.
BIOIMAGING2015-InternationalConferenceonBioimaging
90

3 FOREGROUND ESTIMATION
The three linear filters of the foreground estimation
within the standard image analysis of the beadarray
package can be expressed by means of a single lin-
ear filter. Here, we show the filter equivalent to im-
age sharpening together with the consecutively ap-
plied averaging. We propose its more efficient com-
putation and discuss whether the function meets its
expectations.
Let us denote the intensity in pixel with coordi-
nates [i, j] by w
ij
. The sharpening procedure replaces
w
ij
by
3w
ij
−
w
i, j+1
2
−
w
i, j−1
2
−
w
i+1, j
2
−
w
i−1, j
2
. (1)
In the current code, sharpening is computed over
the whole image and then the foreground intensities
are computed as the weighted combination of four av-
eraged values. These two filters can be namely eas-
ily described by one procedure, saving much compu-
tational complexity in terms of floating point opera-
tions.
This computational improvement based on com-
bining the computation of sharpening and averaging
will be now proposed. It can be easily derived that
both filters can be joined to one replacing w
ij
directly
by linear combination of raw intensities of pixels in
the neighborhood of the pixel [i, j] with coefficients
given by this convolution mask:
0 −1/18 −1/18 −1/18 0
−1/18 2/9 3/18 2/9 −1/18
−1/18 3/18 1/9 3/18 −1/18
−1/18 2/9 3/18 2/9 −1/18
0 −1/18 −1/18 −1/18 0
Besides, there is no justified reason for prefer-
ing this sharpening approach rather than any other
one, e.g. that of (Tanaka et al., 2010), or for com-
bining sharpening with smoothing, because sharpen-
ing increases contrast and smoothing removes it. The
motivation for using sharpening, although formulated
in a vague way, was formulated by (Dunning et al.,
2007) as follows. A bead with a high intensity was
claimed to deplete all of the target molecules from the
neighborhood and therefore the measured intensity is
smaller than the real intensity. A reasonable transfor-
mation should take this into account by increasing the
measured intensity. Such idea is definitely not ful-
filled by the sharpening of formula (1). Actually, (1)
increases a local contrast, but it does not increase the
intensity in a pixel with a large intensity surrounded
by neighbors also with a large intensity. Besides, we
can say that sharpening is accompanied by a conse-
quent averaging, which has the function to improve
the bias introduced by sharpening.
4 ROBUST IMAGE ANALYSIS
The sequence of standard low-level preprocessing in
package beadarray is sensitive to heavy contamina-
tion by a serious noise, which is omnipresent in gene
expression measurements. Therefore, it is highly de-
sirable to replace standard methods by robust coun-
teparts. Tailor-made methods for microarray image
analysis should be proposed using the knowledge
about the causes of noise and errors in the images.
These can either detect outliers and completely ignore
all of them, but other possibilities include to replace
them by values estimated in a suitable model or to
down-weight them.
The standard analysis of BeadChip microarrays
does not exploit available image denoising methods.
On the other hand, denoising itself is complicated and
its numerous methods rely on certain assumptions,
e.g. Gaussian distribution of noise, which may be un-
suitable and in fact allows to preserve some sorts of
outliers. We actually do not have a good experience
if the microarray image analysis is started by a simple
denoising and continues with standard methods.
It is important to robustify all steps of the ap-
proach implemented in beadarray. For example, re-
placing the average by a robust estimator of location
with a high breakdown point turns out to be useful
in estimating the foreground. However, we are not
aware of any application of highly robust methods in
the analysis of microarray images, which would be
based on the concept of robust image analysis. This
has been described as a branch of image analysis ex-
ploiting the ideas and methods of robust statistics with
a high breakdown point.
We are working on implementation of a robust
alternative to the standard approach in C++. Let us
present some key ideas for improving the robustness:
• Robustify each individual step in the whole pro-
cess of the image analysis by a method with a high
breakdown point.
• Replace linear filters by robust counterparts. For
example, flters based on order statistics are more
suitable for impulsive noise.
• Ignore pixels with a nonsense intensity (e.g. zero
intensity).
• Ignore such beads, which have outliers in their
neighborhood. Further, outliers should be re-
moved after each step of the analysis. Only this
can prevent from masking of outliers.
• Treat very bright beads and their direct neighbors
(Figure 1) in a specific way, e.g. delete them or
model their effect and subtract it from the mea-
sured intensities.
RobustImageAnalysisofBeadChipMicroarrays
91

• Perform an artifact detection and correction tailor-
made for artifacts of various sizes and shapes.
• Use a global model for the trend across the whole
strip and use an efficient algorithm for this inten-
sive computation.
To give an example, let us consider estimating the
local background effect. Here, we propose to replace
the standard approach (see Section 2) by the least
weighted squares estimator of location (Kalina, 2012)
computed from w
(6)
,...w
(10)
, where
w
(1)
≤ w
(2)
≤ · · · ≤ w
(25)
(2)
are orderedgray intensities of a circular neighborhood
of a given pixel. Better than that, considering a circu-
lar neighborhood would ensure a rotation invariance
of the computation.
Figure 1: A cut-out of a raw BeadChip microarray im-
age. Its standard analysis does not have a special treatmeant
for highly expressed beads transmitting their signal to their
neighborhood.
Currently, we are comparing various alternative
methods, which are also accompanied by a robust sta-
tistical analysis, using e.g. following ideas.
• Use a robust regularized classification analysis
method, which is suitable for high-dimensional
data, or a robust dimensionality reduction, using
e.g. recently proposed methods of (Filzmoser and
Todorov, 2011; Kalina, 2014).
• Use a weighted classification analysis, where each
gene obtains a weight according to its variability
over beads.
• Classification over beads instead of over gene
transcripts, at the cost of a higher computational
intensity.
• Optimize parameters of image analysis proce-
dures to improve classification results, e.g. by ro-
bust optimization (Xanthopoulos et al., 2013).
5 EXAMPLE
The harmful influence of noise on standard proce-
dures of microarray image analysis will be ilustrated
on real data. We analyzed a data subset from a ge-
netic study of the Center for Biomedical Informatics
(Kalina and Zv´arov´a, 2013). Blood samples of 24 pa-
tients with cerebrovascular stroke and of 24 control
individuals were examined by HumanWG-6 Expres-
sion BeadChips according to manufacturer’s protocol.
First, we investigate the effect of the sharpening
and averaging within the process of foreground esti-
mation. In practice, the user of beadarray may select
to turn it off, while it is not possible to turn off the
averaging. Users are adviced to use sharpening (Dun-
ning et al., 2007; Dunning et al., 2008), although its
influence on the result is extremely high.
We analyzed 48 images (strips of size 2389 ×
18309 pixels) from different microarrays by means
of different approaches, including the option not to
use sharpening and averaging. Figures 2 and 3 show
averaged gene expressions across the 2389 rows of
a microarray strip across 48 microarrays for a partic-
ular gene. The gene was selected randomly as a typi-
cal gene without a differential expression between pa-
tients and control individuals. The plots contain box-
plots of four groups, corresponding to top, upper mid-
dle, bottom middle, and bottom row, respectively. In
other words, the total number of 2389 rows was di-
vided to 4 equally sized groups, while outliers were
discarded from the figures.
Figure 2 shows average foreground intensities
computed without performing sharpening and averag-
ing. Figure 3 analogous values computed with sharp-
ening and averaging. The effect of these transforma-
tions can be seen as a deformation of values in the
last rows of the strip. The figures quantify our ex-
perience with heterogeneity of expressions across the
strip, which we have observed in the majority of pre-
processed (but not raw) microarrays.
Moreover, beadarray detects 0.9 % of beads to be
outliers in the outlier deletion step of the algorithm.
Compared to this strikingly small percentage, our pre-
liminary version of robust image processing considers
as much as 15 % of beads to be too influenced by spa-
tial artifacts or noise and treats them in a specific way.
To quantify the adherse effect of outliers in the
images, we evaluated a simple robust version of the
image preprocessing. Its parameters were tuned to
detect a given percentage of outliers, namely 10 %
and 20 % of measurements (gene expressions) are dis-
carded from the consequent computations.
Further, highly robust principal component analy-
sis LWS-PCA of (Kalina, 2012) was computed from
BIOIMAGING2015-InternationalConferenceonBioimaging
92
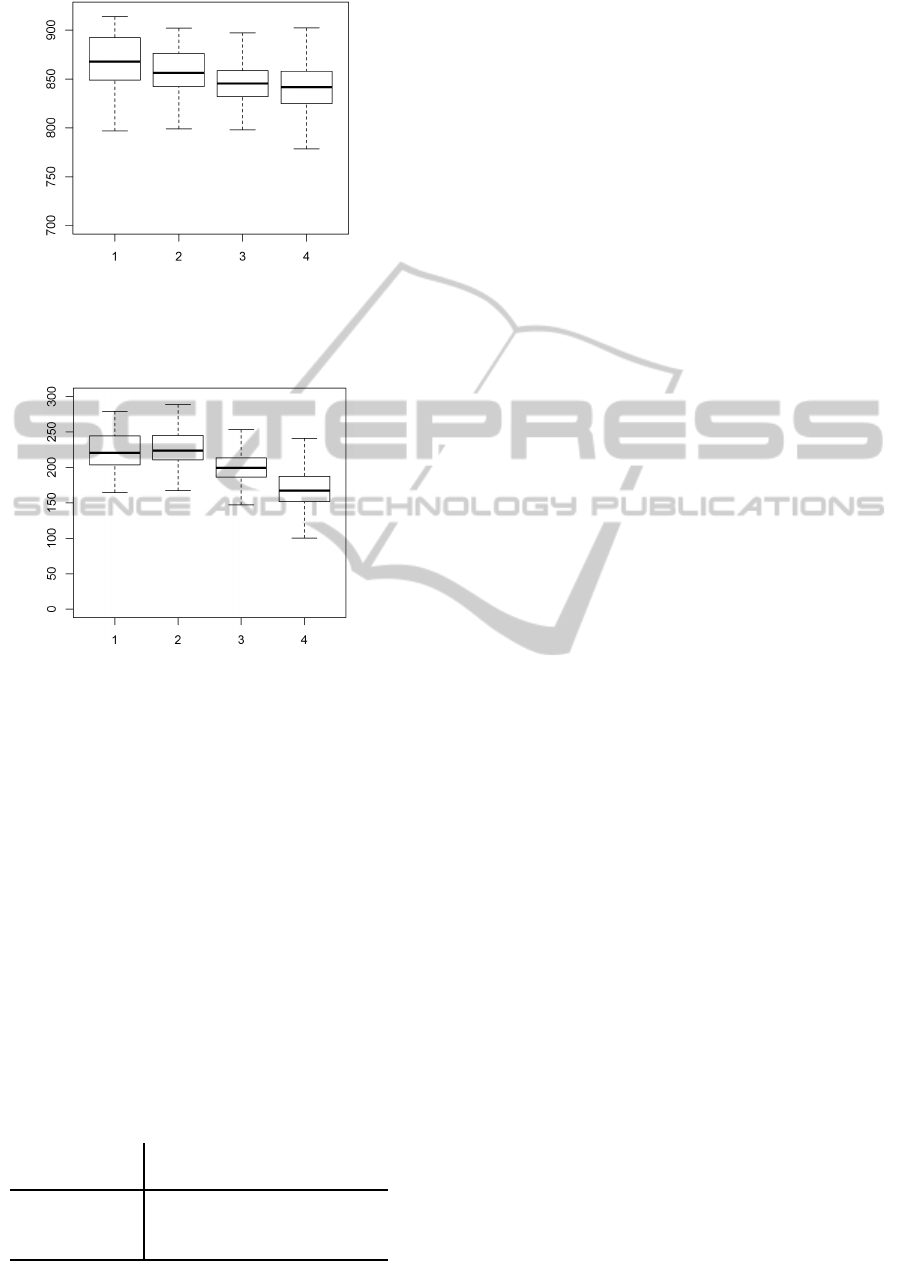
Figure 2: Example of Section 5: Estimated foreground in-
tensities across 48 samples for a particular gene transcript
without performing sharpening and averaging. The image
shows averaged values for top, upper middle, bottom mid-
dle, and bottom rows of the strips.
Figure 3: Estimated foreground intensities across 48 sam-
ples for a particular gene transcript with sharpening and av-
eraging. This is exactly the result of the beadarray package
with default settings of the parameters.
the data. The first row corresponds to the standard ap-
proach. The next rows included trimming off 10 %
and 20 % of measurements, i.e. the fixed number
equal to 10 % (or 20 % of beads are claimed to be
outliers). An increase in the values in Table 1 shows
that the major principal components perform better
in extracting information from the data. In the stan-
dard approach, less important principal components
are namely caused by a mere noise, but their influence
is now reduced closer to zero and the methods can be
interpreted as a denoising (Tibshirani et al., 2003).
Table 1: The contribution (in %) of the major 5 robust prin-
cipal components to explaining the variability of the gene
expression data for various percentage of outliers trimmed
off.
Percentage of
Index
outliers 1 2 3 4 5
0.9 % 6.4 6.1 5.0 4.3 3.9
10 % 7.8 7.4 5.9 5.2 4.0
20 %
8.8 7.8 6.3 5.4 4.1
6 CONCLUSIONS AND FUTURE
RESEARCH
Microarrays remain to be a perspective technology for
research in molecular genetics (G¨ohlmann and Tal-
loen, 2009) and their role is belived to be still in-
creasing (Rueda, 2014). We analyzed the code of
commonly used methods of the beadarray package in
C++, because their details (e.g. the formula (1)) have
not been critically described in literature. This might
have contributed to the fact that sufficient attention
has not been paid to robustness properties of the stan-
dard methodology, not even in monographs on image
analysis of microarrays (Rueda, 2014). This paper re-
veals some disadvantages of the standard image anal-
ysis of BeadChip microarrays.
We promote our position that the standard soft-
ware for the image analysis of BeadChip microar-
rays, which is implemented in the beadarray pack-
age of Bioconductor, suffers from the presence of se-
vere noise in the scanned images. Therefore, we see
a need for software based on an alternative approach
for the BeadChip image analysis. Our criticism of the
standard approaches illustrated on examples with real
data goes far beyond the mild review of (Smith et al.,
2010). The problem is that results of the standard
analysis are biased even on optimally prepared mi-
croarrays. As a consequence, we give a warning that
results of all microarray genetic studies analyzed by
the standard methodology should be interpreted care-
fully, because they are influenced by the high sensi-
tivity of standard procedures with respect to the om-
nipresent contamination of data by noise and outlying
values.
Currently, we are implementing a robust approach
and tune its parameters. Such approach allows a spe-
cific treatment of outliers, spatial artifacts, and global
trend across the strip of the microarray. In this con-
text, pixels influenced by artifacts or severe noise do
not have to be deleted, but carefully modeled, allow-
ing to suppress the effect of noise. After having de-
scribed some principles of robust image analysis of
BeadChip microarrays in this paper, we propose a list
of ideas or topics for a future research in this area.
• Measurement errors can be estimated as the vari-
ability of measured expression in beads with no
corresponding targets in the genome.
• The type of gene probes or the precise location
of beads may be identified wrongly with a rela-
tively large probability, particularly for genes with
a small expression.
• Beads corresponding to highly expressed genes
have a tendency to transmit a part of their signal to
RobustImageAnalysisofBeadChipMicroarrays
93

their neighborhood. Thus, beads with a small in-
tensity may be overilluminated by a strong effect
from their neighborhood (Figure 1).
• Errors in the process of bead localization on the
microarray. They are typical for all beads, but
larger for beads with a lower expression level.
• Errors in the process of bead type identification.
• Urealistic assumption that the expressions of each
gene have the same variability.
• Poisson distribution is more adequate for model-
ing the noise because the measured fluorescence
intensities actually correspond to counts of indi-
vidual photons.
• Sensitivity to discretization of coordinates. The
result is highly influenced by fractional coordi-
nates of the bead, because the assumption of lin-
earity of the intensities of the image between
neighboring pixels is strongly violated.
ACKNOWLEDGEMENTS
The work was financially supported by the Neuron
Foundation for Supporting Science. The work of
A. Schlenker was also supported by the specific re-
search project 260034 (Semantic Interoperability in
Biomedicine and Health Care) of Charles University
in Prague.
REFERENCES
Cardona, A. and Tomancak, P. (2012). Current challenges in
open-source bioimage informatics. Nature Methods,
9:661–665.
Davies, P. and Gather, U. (2005). Breakdown and groups.
Annals of Statistics, 33:997–1035.
Dunning, M., Barbosa-Morais, N., Lynch, A., Tavar´e, S.,
and Ritchie, M. (2008). Statistical issues in the analy-
sis of Illumina data. BMC Bioinformatics, 9(85).
Dunning, M., M.L. Smith, M. R., and Tavar´e, S. (2007).
Beadarray: R classes and methods for Illumina bead-
based data. Bioinformatics, 23:2183–2184.
Filzmoser, P. and Todorov, V. (2011). Review of robust mul-
tivariate statistical methods in high dimension. Ana-
lytica Chinica Acta, 705:2–14.
Fraser, K., Wang, Z., and Liu, X. (2010). Microarray im-
age analysis: An algorithmic approach. Chapman &
Hall/CRC, Boca Raton.
G¨ohlmann, H. and Talloen, W. (2009). Gene expression
studies using Affymetrix microarrays. Chapman &
Hall/CRC, Boca Raton.
Kalina, J. (2012). Implicitly weighted methods in robust
image analysis. Journal of Mathematical Imaging and
Vision, 44:449–462.
Kalina, J. (2014). Classification analysis methods for
high-dimensional genetic data. Biocybernetics and
Biomedical Engineering, 34:10–18.
Kalina, J. and Zv´arov´a, J. (2013). Decision support systems
in the process of improving patient safety. In E-health
Technologies and Improving Patient Safety: Exploring
Organizational Factors. IGI Global, Hershey, 71–83.
Kuhn, K., Baker, S., Chudin, E., Lieu, M.-H., Oeser, S.,
Bennett, H., Rigault, P., Barker, D., McDaniel, T., and
Chee, M. (2004). A novel, high-performance random
array platform for quantitative gene expression profil-
ing. Genome Research, 14:2347–2356.
Rueda, L. (2014). Microarray image and data analysis:
Theory and practice. CRC Press, Boca Raton.
Shi, W., Banerjee, A., Ritchie, M., Gerondakis, S., and
Smyth, G. (2009). Illumina WG-6 beadchip strips
should be normalized separately. BMC Bioinformat-
ics, 10(372).
Smith, M., Dunning, M., Tavar´e, S., and Lynch, A. (2010).
Identification and correction of previously reported
spatial phenomena using raw Illumina BeadArray
data. BMC Bioinformatics, 11(208).
Tanaka, G., Suetake, N., and Uchino, E. (2010). Im-
age enhancement based on nonlinear smoothing and
sharpening for noisy images. Journal of Advanced
Computational Intelligence and Intelligent Informat-
ics, 14:200–207.
Tibshirani, R., Hastie, T., and Narasimhan, B. (2003). Class
prediction by nearest shrunken centroids, with ap-
plications to DNA microarrays. Statistical Science,
18:104–117.
Xanthopoulos, P., Pardalos, P., and Trafalis, T. (2013). Ro-
bust data mining. Springer, New York.
BIOIMAGING2015-InternationalConferenceonBioimaging
94
