
Crutchfield Information Metric for Quantifying the Inter-sequence
Relationship of Multiparametric MRI Data
Jens Kleesiek
1,2,3
, Armin Biller
1,3
and Kai Ueltzh
¨
offer
1
1
Division of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany
2
HCI/IWR, Heidelberg University, Heidelberg, Germany
3
Division of Radiology, German Cancer Research Center, Heidelberg, Germany
Keywords:
Multiparametric MRI, Crutchfield Information Metric, MRI Quality Control.
Abstract:
A plethora of different MRI sequences exists. To automatically structure this ’zoo’ of available sequences we
propose the usage of a framework rooted in information theory. In this paper we show that the Crutchfield
information metric is a suitable distance measure for this purpose. It is demonstrated that the physical relation-
ship can be inferred with this metric solely based on the voxel intensities. As future applications we envisage
MRI sequence quality control and standardization.
1 INTRODUCTION
The necessity of multiparametric magnetic resonance
imaging (MRI) for diagnosis and therapy monitoring
of diseases is undoubted. However, in general there
are no standardized protocols specified (Cornfeld and
Sprenkle, 2013). In the clinical routine workup it is
not feasible to acquire every available sequence de-
posited at the scanner. This would result in an unrea-
sonable long scanning time, very likely making the
patient feel uncomfortable. Secondly, the images ac-
quired at the end of a long scanning session tend to
suffer from motion artifacts which impair their diag-
nostic value. Needless to say that this approach is eco-
nomically unacceptable.
To shed light on the zoo of available MRI se-
quences we try to establish a framework that is rooted
in information theory and allows us to capture and
quantify the relative information content between
MRI sequences. The current work should be seen as
a proof of concept which in the future can be an aid
for standardizing MRI protocols and possibly also be
used to optimize MRI sequence parameters.
Historically, information theory investigated the
transmission between a sender and a receiver (Shan-
non and Weaver, 1949) but it has also been extended
to theoretical measures that capture the information
integration (Tononi et al., 1998) and information dis-
tances of information sources (Kullback, 1968). A
not well known but still very important information
distance was introduced by Crutchfield (Crutchfield,
1990). He showed that a proper metric space (in
a mathematical sense) of information sources can
be defined. Given physically or functionally related
sources, i.e. different MRI sequences, that are acti-
vated by an identical localized stimulus, in our case a
patient that is examined, the information distance be-
tween those sources can be determined using this met-
ric. In turn, due to the fact that it is a proper metric we
can exploit the discovered relationship geometrically.
To the best of our knowledge no comparable ap-
proach has been examined for multiparametric MRI
data previously. We were inspired by robotic exper-
iments were the Crutchfield information metric has
been used to determine the informational topology
of a set of robot sensors, that consecutively was ex-
ploited for simple visually guided movements (Ols-
son et al., 2006) or unsupervised activity classifica-
tion (Kaplan and Hafner, 2006).
2 MATERIALS AND METHODS
2.1 Theory
Each MRI sequence can be interpreted as an infor-
mation source with the voxel intensities as the re-
spective measurements. Given two different informa-
tion sources X and Y , e.g. corresponding T1- and T2-
weighted data sets, it is possible to compute the con-
ditional entropy of the two sources:
5
Kleesiek J., Biller A. and Ueltzhöffer K..
Crutchfield Information Metric for Quantifying the Inter-sequence Relationship of Multiparametric MRI Data.
DOI: 10.5220/0005181800050012
In Proceedings of the International Conference on Bioimaging (BIOIMAGING-2015), pages 5-12
ISBN: 978-989-758-072-7
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

H(X|Y ) = −
∑
x
∑
y
p(x, y) log
2
p(x|y) . (1)
Consecutively, given H(X |Y ) and the entropy
H(X) allows to determine the joint entropy:
H(X, Y ) = H(X) + H(Y |X)
=
∑
x
p(x)log
2
p(x) + H(Y |X) .
(2)
After calculating these quantities the distance be-
tween the two information sources can be com-
puted in the form of the Crutchfield information met-
ric (Crutchfield, 1990):
d
C
(X, Y ) =
H(Y |X) + H(X|Y )
H(X, Y )
. (3)
This metric is related to the mutual information
(MI). However, MI measures what two random vari-
ables have in common, whereas the Crutchfield in-
formation metric quantifies what they do not have in
common (Olsson et al., 2006). In Addition, in con-
trast to the MI the Crutchfield distance is a proper
metric fullfilling the properties of:
1. symmetry: d
C
(X, Y ) = d
C
(Y, X),
2. equivalence: d
C
(X, Y ) = 0 iff X and Y
are recoding-equivalent (as defined by Crutch-
field (Crutchfield, 1990)), d
C
(X, Y ) = 1 states that
the two sources are independent,
3. triangle inequality: d
C
(X, Z) ≤ d
C
(X, Y ) +
d
C
(Y, Z).
Being a metric implies that the information space
has a structure that can be exploited geometrically.
The first experiment is a proof of concept. In the
second experiment we computed the metric for all
combinations (D = 171) of sequences within a pri-
vate multiparametric MRI data set (see Sec. 2.2) and
interpreted the resulting matrix as a distance (dis-
similarity) matrix and as a graph adjacency matrix.
In the former case we used non-linear dimensional-
ity reduction methods like Isomap (Tenenbaum et al.,
2000) and local linear embedding (Roweis and Saul,
2000) to embed the data in a 2D geometric space, in
the latter case we used Kruskal’s algorithm (Kruskal,
1956) to obtain the minimum spanning tree (MST). In
a third experiment we computed the distance matrices
(N = 216, D = 6) for a publicly available multipara-
metric MRI data set (see Sec. 2.2) and used the low
dimensional embedding to identify impaired images.
2.2 Data
2.2.1 Data Set 1
In total N = 17 multiparametric MRI data sets with
C = 19 channels each were acquired in a clinical rou-
tine workup of patients using two different 3 Tesla
MR system from the same manufacturer (Magne-
tom Tim Trio (N = 9) and Magnetom Verio (N = 8),
Siemens Healthcare, Erlangen, Germany).
All patients were suffering from a glioblas-
toma multiforme (WHO grade IV). The data was
anonymized. The reasoning for taken data from
diseased subjects is motivated by the fact that pa-
tients suffering from this disease are scanned with a
more detailed protocol that comprises more MRI se-
quences.
The following MRI images were acquired: na-
tive (T1) and contrast enhanced (T1CE) T1-weighted
images with T E = 4.04 ms and T R = 1710 ms; T2-
weighted TSE imaging (T2) with T E = 85 ms and
T R = 5500 ms; T2-weighted fluid attenuated inver-
sion recovery images (FLAIR) with T E = 135 ms
and T R = 8500 ms; diffusion-weighted images with
T E = 90 ms and T R = 5300 ms comprising a b =
0 (DWI b0), a b = 1200 (DWI b1200t) as well as
an apparent diffusion coefficient (ADC) map; native
(SWI) and contrast enhanced (SWICE) susceptibil-
ity weighted images with T E = 19.7 ms and T R =
27 ms, this set of sequences also includes a mag-
nitude (SWI[CE] MAG) and phase (SWI[CE] PHA)
image as well as a minimum intensity projection
(SWI[CE] MIP); dynamic susceptbility contrast per-
fusion images with T E = 37 ms and T R = 2220 ms
yielding the relative cerebral blood flow (PWI CBF)
and volume (PWI CBV), the mean transit time
(PWI MTT) and the time to peak (PWI TTP).
2.2.2 Data Set 2
For the third experiment we use the publicly available
BraTS 2014 training data set provided via the Vir-
tual Skeleton Database (VSD) (Kistler et al., 2013).
It comprises N = 216 co-registered native and con-
trast enhanced T1-weighted images, as well as T2-
weighted and T2-FLAIR images (C = 4). The data
was acquired with MR scanners of different vendors,
at different field strengths and using non-uniform pro-
tocols (i.e. physical parameters). The images contain
low grade as well as high grade tumors.
BIOIMAGING2015-InternationalConferenceonBioimaging
6

2.3 Preprocessing
2.3.1 Experiment 1 and 2
All sequences of the multiparametric data set 1 were
co-registered intra-individually to the respective na-
tive T1-weighted images. A rigid 6-DOF registration
was preformed using the BRAINSFit (Johnson et al.,
2007) command line interface of 3D-Slicer (3D Slicer
v4.3). The registration accuracy was confirmed by a
board-certified neuroradiologist. In the next step we
used FMRIB’s brain extraction tool (BET) (Smith,
2002), which is part of FSL (FMRIBs Software Li-
brary FSL v5.0), for deskulling of the T1-weighted
images. The obtained mask was applied to all chan-
nels. Finally, all data was rescaled to be in the range
[0, 1024]. To estimate the probabilities we used a stan-
dard frequency count method, after we confirmed that
histogram equalization methods do not alter the re-
sults.
2.3.2 Experiment 3
Data set 2 was rescaled to be in the range [0, 1024]
and the same standard frequency count method as in
the first two experiments was applied.
2.4 Data Manipulation
For the first experiment we manipulated the data. We
added increasing levels of noise to the images using a
normal distribution N (µ = 0, σ
2
) centered at zero but
with varying values of sigma. Further, we generated
an artificial sequence Z by combining two recorded
sequences X and Y with a weighted sum:
Z = (1 − α)X +αY . (4)
Finally, we applied a 6-DOF rigid transformation T
that allows for separately rotating around an axis or
translating along an axis.
2.5 Implementation
Except the two tools (BRAINSFit (Johnson et al.,
2007) and BET (Smith, 2002)) noted above, all
algorithms were implemented in custom python
scripts (Python v2.7.6). For analyzing the distance
matrices we adapted functions implemented in scikit-
learn (Pedregosa et al., 2011) and NetworkX (Hag-
berg et al., 2008).
3 RESULTS
3.1 Experiment 1 – Proof of Concept
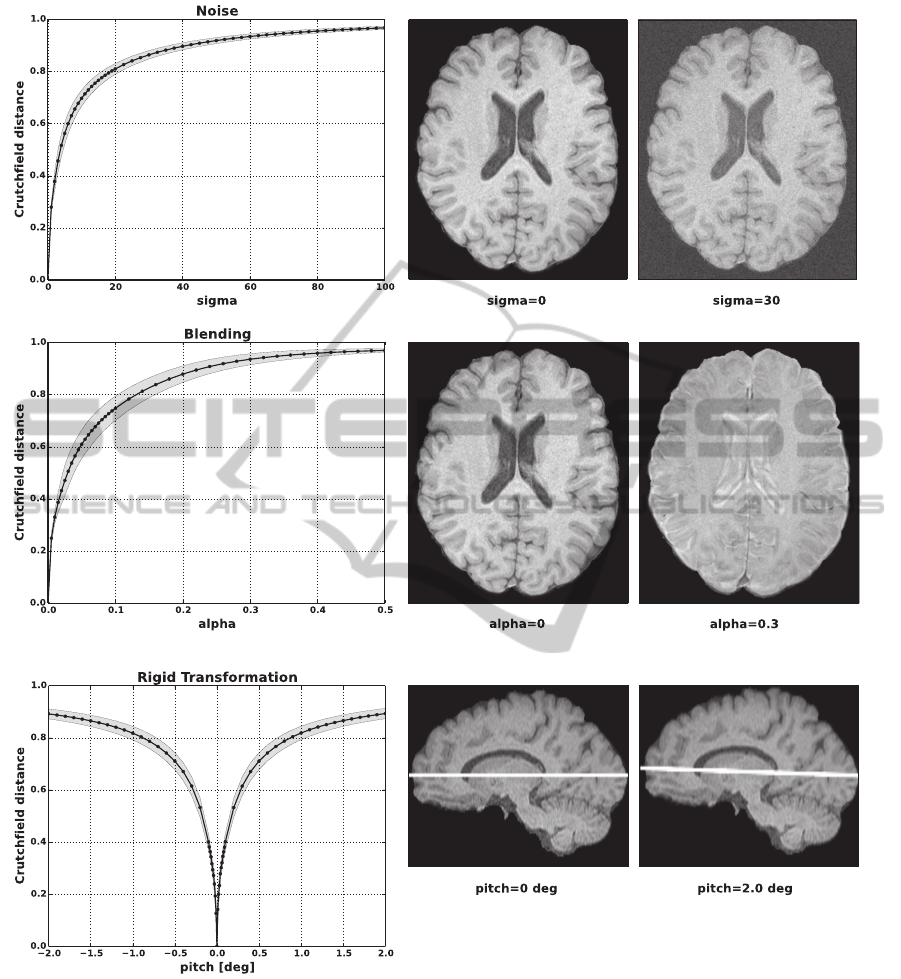
In the first experiment (Fig. 1) we confirmed that the
Crutchfield information distance is a valid metric for
our purposes. On this account we manipulated the
MRI image data (data set 1) in several ways. The av-
erage course as well as the standard deviation (gray
shaded area) are plotted for all experiments.
Initially we added an increasing level of gaussian
noise to the data. This manipulation was repeated
for each T1 sequence of MRI data set 1 (N = 17).
In Fig. 1A left it can be seen that the Crutchfield
distance increases monotonically with the amount of
noise added. In Fig. 1A right an exemplary axial T1-
weighted image is shown without noise and with a
noise level corresponding to σ = 30.
Secondly, we generated artificial MRI data by
blending the T1-weighted and T2-weighted images of
the same, co-registered data set using varying weight-
ing factors (Eq. 4). We then computed the Crutch-
field distance between the T1 sequence and the artifi-
cial images. This was repeated for all images of MRI
data set 1. In Fig. 1B left it can be seen that, as ex-
pected, with an increasing T2-fraction of the artificial
image also the Crutchfield distance to the T1 image
increases. In Fig. 1B right an exemplary axial T1 slice
as well as a mixed T1- and T2-weighted image using
a factor of α = 0.3 are shown.
Next, we used a rigid transformation to selectively
rotate a data set around an axis. We measured the
Crutchfield distance of an unmodified reference se-
quence to the identical, but rotated data set in the in-
terval of [−2, 2] degrees. This is shown for the pitch
movement in Fig. 1C left. It clearly can be seen that
there is a well defined minimum at 0 degrees with an
almost symmetrically increasing information distance
in both rotation directions. As an example, an unro-
tated sagittal T1-weighted image as well as an image
pitched by 2 degrees is presented in Fig. 1C right.
3.2 Experiment 2 – Application to
Multiparametric MRI Data
Fig. 2A shows the Crutchfield information distance
for all combinations (D = 171) of sequences from
data set 1. Note the symmetry of the matrix. We
depicted the average of all N = 17 multiparametric
MRI data sets. However, the structure that can be
seen is also present at the individual level. This is
also supported by the small standard deviation of the
distances, which is on average 0.0035.
CrutchfieldInformationMetricforQuantifyingtheInter-sequenceRelationshipofMultiparametricMRIData
7
A
B
C
Figure 1: Data manipulation. A) Increasing levels of gaussian noise were added to a T1-weighted image and the Crutchfield
distance to the original sequence was determined (left). Exemplary axial T1 image with (σ = 30) and without noise (right). B)
Crutchfield distance between a T1-weighted image and artificially generated images (left). The artificial images were obtained
by blending co-registered T1- and T2-weighted images. Exemplary axial T1 image as well as a mixed T1- and T2-weighted
image using a factor of α = 0.3 (right). C) Crutchfield distance of an unmodified reference sequence to the identical, but in
the interval of [−2, 2] degrees rotated data set (left). Unrotated sagittal T1 image as well as an image pitched by 2 degrees
(right). The average course for all data sets N = 17 as well as the standard deviation (gray shaded area) are plotted.
First, we interpreted the distance matrix as a
dis-similarity matrix. We used the Isomap algo-
rithm (Tenenbaum et al., 2000) to perform a 2D em-
bedding of the data (Fig. 2B). Comparable results
were obtained, when we employed local linear em-
bedding (Roweis and Saul, 2000) to reduce the di-
mensionality of the data (not shown). It clearly can
be seen that related sequences cluster in close proxim-
BIOIMAGING2015-InternationalConferenceonBioimaging
8
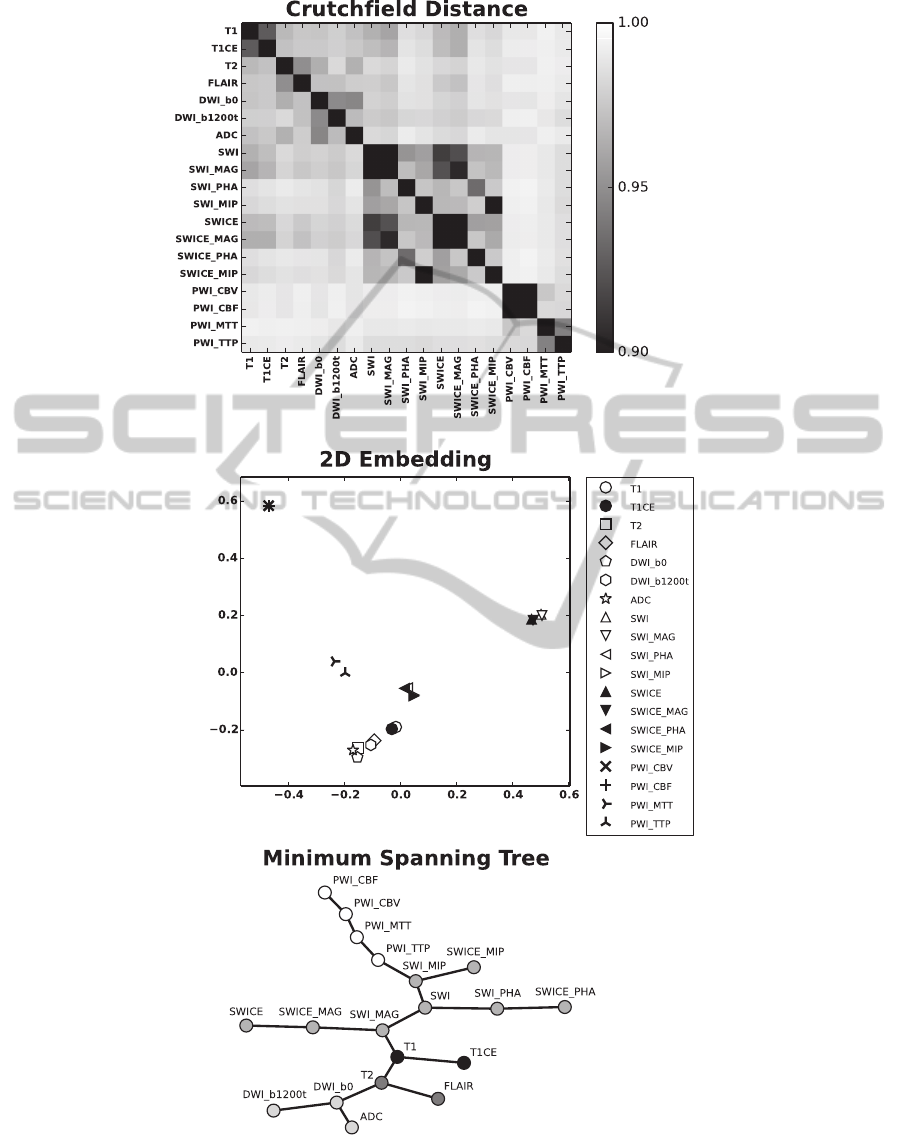
A
B
C
Figure 2: Crutchfield Matrix. A) Average Crutchfield information distance between all combinations of the multiparametric
MRI data of data set 1. The scaling ([0.9, 1.0]) was chosen to better emphasize the structure. B) 2D geometric embedding of
the distance matrix depicted in A using the Isomap algorithm. C) MST computed from the distance matrix depicted in A using
Kruskal’s algorithm. For the embedding we used the graphviz “spring model” layout (Graphviz). For the abbreviations of the
MRI sequences please refer to Sec. 2.2.
CrutchfieldInformationMetricforQuantifyingtheInter-sequenceRelationshipofMultiparametricMRIData
9

ity. In a second approach we used the distance matrix
as an adjacency matrix of a fully connected graph and
applied Kruskal’s algorithm (Kruskal, 1956) to obtain
the MST (Fig. 2C). Also this method allows to dis-
cover physically related sequences by grouping them
at neighboring leaves in the tree.
3.3 Experiment 3 – Automatic Quality
Control
To demonstrate a potential application of the pro-
posed framework we compute the distance matri-
ces for N = 216 data sets from the BraTS 2014
training data (data set 2) and use the Isomap algo-
rithm (Tenenbaum et al., 2000) to perform a 2D em-
bedding (Fig. 3A). Based on the distance to the clus-
ter centers we were able to identify outliers. This is
shown for four cases (numbered 1 to 4 in Fig. 3A). To
confirm our hypothesis that the outliers correspond to
impaired data sets that do not meet quality standards,
we manually inspected them. Case 1 corresponds to
data set brats tcia pat313 1. This data set contains
no native T1 - instead the T2-FLAIR image was en-
closed twice. Case 2 (brats tcia pat216 1) misses
again a native T1 weighted image. Instead a contrast
enhanced image with spherical artifacts was included
(Fig. 3B left). Case 3 (brats tcia pat230 2) displays
severe motion artifacts in T1 (Fig. 3B right). Case 4
corresponds to brats tcia pat250 1 and does not con-
tain a native T1, instead a T1CE was included twice.
Note, even if only one channel is corrupted this
leads to changes of multiple entries in the distance
matrix and thus can affect the position of (all) other
channels in the low dimensional embedding.
4 DISCUSSION
We present an information theoretic framework that
allows to infer the relationship of MRI sequences
purely based on voxel intensities. It is shown that the
Crutchfield information metric (Crutchfield, 1990) is
a suitable distance measure for MRI sequences and
is able to capture the following relationship: the
greater the (physical) distance between two MRI se-
quences, the less information they share. We ma-
nipulated images by adding noise (Fig. 1A), blending
two MRI sequences (Fig. 1B) and purposefully apply-
ing a rigid transform to them (Fig. 1C). In all cases
the Crutchfield distance increased monotonically with
the amount of manipulation and showed only a small
standard deviation across data set 1 (N = 17). If we
measure the information distance between all com-
binations of sequences (D = 171) of data set 1, we
can construct a distance matrix which already shows
a structure that corresponds to the intrinsic physical
relationship of the sequences (Fig. 2A). This relation-
ship becomes more explicit if we perform a low di-
mensional (2D) embedding (Fig. 2B) or compute the
MST (Fig. 2C).
Usually the physical relationship of the MRI se-
quences is known or can be obtained from the DI-
COM header. What is the benefit of the proposed
method? This objection is certainly valid, however,
consider for instance data from a multicenter study
which is designated for an automatic evaluation. Even
if the data sets are acquired with similar parameters
(e.g. TE and TR) they still originate from different
scanners and thus might not be located in the same
informational space. It also is very likely, as known
from clinical routine, that some of the channels are
affected by motion artifacts, which would also al-
ter the informational structure. We demonstrated for
N = 216 data sets that the proposed framework in-
deed can be used as an automated screening method
for impaired images (Fig. 3). Employing the Crutch-
field metric for quality control allows to identify data
sets which are not located in the same informational
space, e.g. are affected by motion artifacts (Fig. 3B
right). Admittedly, so far this is a very coarse ap-
proach and it still has to be validated on a finer level
with controlled experiments that determine sensitivity
and specificity of the method.
Another potential application is to utilize this
method for the assembly of standardized multipara-
metric MRI sequences. The information distance can
be used as guideline for radiologists to select opti-
mal subsets of the available sequences by e.g. prun-
ing the MST to minimize the aquisition of redundant
information. Further applications include MRI se-
quence optimization by choosing parameters of a set
of sequences to maximize the coverage in information
space, i.e. reducing redundancy within the sequences.
Yet, this still requires a thourough study of the depen-
dence of the Crutchfield distance on the differences in
physical parameters of MRI sequences.
5 CONCLUSIONS
We demonstrated that the Crutchfield information
metric in combination with methods for dimension-
ality reduction or from graph theory are suitable for
discovering the physical relationship of various MRI
sequences solely based on their voxel intensities. Ini-
tial experiments confirm that the proposed framework
can be used for automatic MRI sequence quality con-
trol. This has to be validated in future work.
BIOIMAGING2015-InternationalConferenceonBioimaging
10
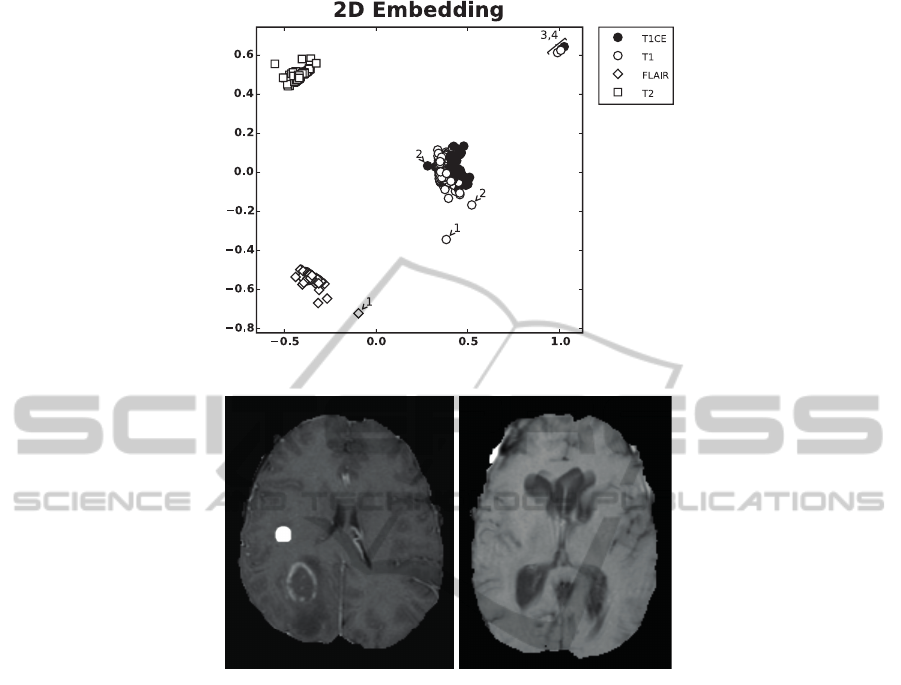
A
B
Figure 3: 2D Embedding of BraTS 2014 training data. A) Using the Isomap algorithm we embedded all N = 216 data sets
in 2D. This allowed us to identify outliers (e.g. numbered 1 to 4) which indeed corresponded to impaired data sets. The scatter
of the embedded points can be explained by the fact that the data was acquired with MRI scanners of different vendors as well
as with different field strengths and protocols. For the abbreviations of the MRI sequences please refer to Sec. 2.2. B) The
left image corresponds to number 2 above and was labeled as a native T1 weighted image. Instead it is a contrast enhanced
T1 image that contains multiple spherical hyperintense artifacts (a neuroradiologist confirmed that these do not correspond to
hemorrhage). The image on the right side exhibits severe motion artifacts and corresponds to number 3 above.
ACKNOWLEDGEMENTS
This work was supported by a postdoctoral fellowship
from the Medical Faculty of the University of Heidel-
berg.
REFERENCES
3D Slicer v4.3. http://www.slicer.org.
Cornfeld, D. and Sprenkle, P. (2013). Multiparametric MRI:
standardizations needed. Oncology (Williston Park),
27(4):277,280.
Crutchfield, J. (1990). Information and its metric. In Non-
linear Structures in Physical Systems – Pattern For-
mation, Chaos and Waves, pages 119–130. Springer
Verlag.
FMRIB’s software Library FSL v5.0.
http://fsl.fmrib.ox.ac.uk.
Graphviz. http://www.graphviz.org.
Hagberg, A. A., Schult, D. A., and Swart, P. J. (2008).
Exploring network structure, dynamics, and function
using NetworkX. In Proceedings of the 7th Python
in Science Conference (SciPy2008), pages 11–15,
Pasadena, CA USA.
Johnson, H., Harris, G., and Williams, K. (2007). BRAINS-
Fit: Mutual Information Registrations of Whole-Brain
3D Images, Using the Insight Toolkit. The Insight
Journal.
Kaplan, F. and Hafner, V. V. (2006). Information-theoretic
framework for unsupervised activity classification.
Advanced Robotics, 20(10):1087–1103.
Kistler, M., Bonaretti, S., Pfahrer, M., Niklaus, R., and
B
¨
uchler, P. (2013). The virtual skeleton database: an
CrutchfieldInformationMetricforQuantifyingtheInter-sequenceRelationshipofMultiparametricMRIData
11

open access repository for biomedical research and
collaboration. J Med Internet Res, 15(11):e245.
Kruskal, Joseph B., J. (1956). On the shortest spanning
subtree of a graph and the traveling salesman problem.
Proceedings of the American Mathematical Society,
7(1):pp. 48–50.
Kullback, S. (1968). Information Theory and Statistics.
Dover, New York.
Olsson, L. A., Nehaniv, C. L., and Polani, D. (2006). From
unknown sensors and actuators to actions grounded
in sensorimotor perceptions. Connection Science,
18(2):121–144.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V., Vanderplas, J., Passos, A.,
Cournapeau, D., Brucher, M., Perrot, M., and Duch-
esnay, E. (2011). Scikit-learn: Machine Learning
in Python. Journal of Machine Learning Research,
12:2825–2830.
Python v2.7.6. http://www.python.org.
Roweis, S. T. and Saul, L. K. (2000). Nonlinear dimension-
ality reduction by locally linear embedding. Science,
290(5500):2323–6.
Shannon, C. and Weaver, W. (1949). The Mathemati-
cal Theory of Communication. University of Illinois
Press, Chicago, IL.
Smith, S. M. (2002). Fast robust automated brain extraction.
Human brain mapping, 17(3):143–55.
Tenenbaum, J. B., de Silva, V., and Langford, J. C. (2000).
A global geometric framework for nonlinear dimen-
sionality reduction. Science, 290(5500):2319–23.
Tononi, G., Edelman, G. M., and Sporns, O. (1998). Com-
plexity and coherency: integrating information in the
brain. Trends Cogn Sci, 2(12):474–84.
BIOIMAGING2015-InternationalConferenceonBioimaging
12
