
A Method for Evaluating Validity of Piecewise-linear Models
Oleg V. Senko
1
, Dmitriy S. Dzyba
2
, Ekaterina A. Pigarova
3
, Liudmila Ya. Rozhinskaya
3
and Anna V. Kuznetsova
4
1
Dorodnicyn Computing Center, Russian Academy of Sciences, ul. Vavilova 40, 119991 Moscow, Russia
2
Lomonosov Moscow State University, Leninskie Gory, Moscow, Russia
3
Department of Neuroendocrinology and Bone Diseases, Endocrinology Research Centre,
11 Dmitry Ulyanov st., 117036 Moscow, Russia
4
Emanuel Institute of Biochemical Physics, ul. Kosygina 4, 117997 Moscow, Russia
Keywords:
Regression Model, Optimal Complexity, Permutation Test.
Abstract:
A method for evaluating optimal complexity of regression models is discussed. It is supposed that complicated
model must be used only when any simple model fails describe exhaustively regularity that exists in data.
At that null hypothesis about exhaustive explanation of data by simple regularity is tested with the help of
complicated model. Validity of null hypothesis is evaluated with the help p-value that is calculated with the
help of special version of permutation test. An application is discussed where developed technique is used
to evaluate if more complicated piecewise-linear regressions must be used instead of simple regressions to
describe correctly dependence of parathyroid hormone on vitamin D status.
1 INTRODUCTION
Standard task of statistical modelling is discussed. It
is necessary to find statistical model that forecasts re-
sponse Y by variables X
1
, . . . , X
n
:
Y = F(X
1
, . . . , X
n
) + ε,
where F(X
1
, . . . , X
n
) is predicting function and ε is
error term. Function F with minimal mathemati-
cal mean Eε
2
is chosen from family
e
M by data set
e
S
0
= {(y
0
1
, x
0
1
), . . . , (y
0
m
, x
0
m
)}, where y
0
1
, . . . , y
0
m
are val-
ues of response variable Y and x
0
1
, . . . , x
0
m
are vectors
of predicting variables X
1
, . . . , X
n
. It is supposed that
observations corresponding different objects from
e
S
0
are independent and are taken from the same proba-
bility space. Success of modelling depends on correct
choice of predicting function F complexity or more
exactly on complexity of family
e
M. Today there are
several approaches for complexity optimization that
allow to discourage overfitting effect. Akaike infor-
mation criterion (Akaike, 1974), Bayesian informa-
tion criterion (Schwarz, 1978), Hannan-Quinn infor-
mation criterion (Hannan and Quinn, 1979), Risannen
principle (Rissanen, 1978) may be mentioned there
above. These techniques often allow to find out com-
plexity level with best generalization ability. But in
many application tasks it is important not only to find
model of optimal complexity but also to estimate va-
lidity of choice. Let suppose that models may be
searched inside simple family
e
M
s
and more compli-
cated family
e
M
c
. At that
e
M
s
⊆
e
M
c
. It is not sufficient
to find out if optimal model must be searched inside
family
e
M
s
or inside family
e
M
c
\
e
M
s
. It is also neces-
sary to evaluate our confidence that model found in-
side
e
M
c
\
e
M
s
really better describes data than model
found inside family
e
M
s
. It must noted that choice be-
tween two families sometimes corresponds to choice
between two suppositions about type of process that
generates studied data. It may be physical, chemical
or biological process for example. Usually in statis-
tics validity of choice between two hypotheses is eval-
uated with the help of p-values. The same way of
validity evaluating is used in this paper. It is consid-
ered that complicated family must be used then and
only then when any simple model fails to describe ex-
haustively regularity that exists in data. At that null
hypothesis about exhaustive explanation of existing
regularity by simple predictive function from
e
M
s
is
tested with the help of complicated family
e
M
c
. Such
approach correspond to well known principle of of
Occam’s razor that is attributed William of Occam
living in the 14th century. The most popular ver-
sion of razor is formulated as ”Entities should not
be multiplied beyond necessity.” Later razor principle
437
V. Senko O., S. Dzyba ., A. Pigarova E., Ya. Rozhinskaya L. and V. Kuznetsova A..
A Method for Evaluating Validity of Piecewise-linear Models.
DOI: 10.5220/0005156904370443
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2014), pages 437-443
ISBN: 978-989-758-048-2
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

was adopted by many scientists and another variants
were invented. Principle was stated by Isaac New-
ton in form ”We are to admit no more causes of nat-
ural things than such as are both true and sufficient
to explain their appearances.” Such form as it may be
seen is most close to approach that is represented in
the paper. Problems that are associated with Occam’s
razor are discussed in modern scientific literature on
machine learning or knowledge discovery. Usually
it is considered that razor is a way to improve fore-
casting ability. Arguments for and against such razors
are represented in details in (Domingos, 1999). Ap-
proach that is discussed in this paper is based on test-
ing of null hypotheses with the help of random per-
mutation test. Let note that random permutation test
now is rather popular technique allowing to evaluate
statistical validity without any additional suppositions
(Ernst, 2004; Good, 2005). Permutation tests also are
used to study regression or recognition models (Kim
et al., 2000; Ojala and Garriga, 2010; Golland et al.,
2000).
2 EVALUATING VALIDITY OF
COMPLICATED MODELS
2.1 Main Suppositions
It is supposed that optimal predicting function F
0
(x) is
searched inside family widetildeM by some training
set
e
S = {(y
1
, x
1
), . . . , (y
m
, x
m
)} with the help of least
squares technique:
F
0
(x) = argmin
F∈
e
M
Q[
e
S, F(x)],
where Q[
e
S, F] =
∑
m
j=1
[y
j
− F(x
j
)]
2
. Minimal value of
Q[
e
S, F(x)] at set
e
M will be referred to as Q
min
(
e
S,
e
M).
The represented approach is based on several simple
suppositions.
Supposition 1. More complicated function from
e
M
c
must be used only when there is no function inside
family
e
M
s
that exhaustively describes data.
Supposition 2. It is considered that some func-
tion F exhaustively describes dependence of Y from
X
1
, . . . , X
n
if residuals {r
1
= y
1
− F(x
0
1
), . . . , r
m
=
y
m
− F(x
0
m
)} are realizations of mutually independent
identically distributed random values ξ
1
, . . . , ξ
m
that
are independent on vector descriptions x. It is sup-
posed also that E(ξ
i
) = 0, i = 1, . . . , m}.
Supposition 3. It is possible to reject (or verify) null
hypothesis that function F exhaustively describes de-
pendence on X variables with the help of complicated
family
e
M
c
.
2.2 Permutation Test Technique
Let
e
f is set of all possible permutations of integers
{1, . . . , m}. Let
e
S
p
( f, F) be data set that is received
from initial data set
e
S
0
by random permutation of
residuals (r
1
, . . . , r
m
):
e
S
p
( f, F) = {[r
f(1)
+F(x
0
1
), x
0
1
], . . . , [r
f(m)
+F(x
0
m
), x
0
m
]}.
Definition Two permutations f
′
and f
′′
from
e
f will be
called equivalent if data sets
e
S
p
( f
′
, F) and
e
S
p
( f
′′
, F)
are equal.
Let
e
f
b
= { f
b
1
, . . . , f
b
N
} is such set of permutations
that
• any two permutation from
e
f
b
are not equivalent,
• any permutation is equivalent to one of permuta-
tions from
e
f
b
.
Let note that due to transitiveness of equivalence any
permutation may be equivalentonly one element from
e
f
b
. Equivalence class c( f) may be defined for each
permutation from
e
f
b
that consists of all permutation
that are equivalent to f. Equality
e
f =
N
[
i=1
c( f
b
j
)
is true by definition of
e
f
b
. Two statement are true.
Statement 1. In case supposition 2 is true for any
f
j
∈
e
f
b
P{
e
S
p
[ f
j
, F] | x
1
= x
0
1
, . . . , x
m
= x
0
m
} =
m
∏
i=1
P(ξ
i
= r
i
)
Proof. Statement 1 may be easily received from
independence of residuals r on vectors x and mutual
independence of observations corresponding different
objects from
e
S
0
. It follows from supposition 2 that
probabilities of data sets
e
S
p
( f
1
, F), . . . ,
e
S
p
[ f
N
, F] are
equal each other. Q.E.D.
Statement 2. All classes c[ f
1
], . . . , c[ f
N
] are of the
same size.
Proof. Really. Let {er
1
, . . . , er
k
} be such partition of
{r(1), . . . , r(m)} that residuals r
∗
inside each ele-
ment of partition are equal each other and residuals
from different groups are different. Suppose that
e
J
q
= {J
q
(1), . . . , J
q
[µ(q)]} is set of residuals numbers
inside group er
q
according some permutation f
j
∈
e
f
b
,
where µ(q) is size of group er
q
and q = 1, . . . , k. It is
evident that for any permutation f
′
j
that is received
from f
j
by some permutations of numbers only inside
sets
e
J
1
, . . . ,
e
J
k
equality of data sets
e
S
p
( f
j
, F) and
e
S
p
( f
′
j
, F) is preserved. At that for any permutation
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
438

f
′′
j
that is received from f
j
by some permutation
including exchanges between sets
e
J
1
, . . . ,
e
J
k
data sets
e
S
p
( f
j
, F) and
e
S
p
( f
′′
j
, F) are not equal. So class c( f
j
)
must include all permutations that are received from
f
j
by some permutations of numbers inside sets
e
J
1
, . . . ,
e
J
k
. Class c( f
j
) does not include any other per-
mutations. Let note that amount of such permutations
depends only on sizes of groups {er
1
, . . . , er
k
} and does
not depend on specific permutation f
j
. So size of
class c[ f
j
] does not depend on f
j
. Q.E.D.
Set
e
S
b
= {
e
S
p
( f
1
, F), . . . ,
e
S
p
( f
N
, F)} includes all
possible data sets
e
S satisfying conditions
a) empirical distribution of residuals r from fore-
casting function F in
e
S coincides with empirical
distribution of residuals r at initial data set
e
S
0
(condi-
tion C
r
(
e
S
0
, F));
b) x-descriptions in
e
S completely coincide with
x-descriptions of
e
S
0
(condition C
x
(
e
S
0
, F)).
Let P is some predicate that is defined at set of all
possible data sets of size m. Let predicate P be true
at some subset
e
S
T
(P ) of set
e
S
b
. Probabilities of all
data sets from
e
S
b
are equal according statement 2. So
equality P{P (
e
S) = TRUE|
e
S ∈
e
S
b
} may be evaluated
as ratio
|
e
S
T
(P )|
|
e
S
b
|
.
Supposition 4. Let P
pv
= Q
min
(
e
S,
e
M
c
) <
Q
min
(
e
S
0
,
e
M
c
). It is suggested to use conditional
probability
P{P (
e
S) = TRUE|
e
S ∈
e
S
b
} =
|
e
S
T
(P
pv
)|
|
e
S
b
|
(1)
as p-value that evaluates validity of null hypothesis
about exhaustiveness.
Statement 3. Equality is true
|
e
S
T
(P
pv
)|
|
e
S
b
|
=
=
| f ∈
e
f|Q
min
[
e
S
p
( f, F),
e
M
c
] < Q
min
(
e
S
0
,
e
M
c
)|
|
e
f|
(2)
Proof. Really, evidently
P ]
e
S
p
( f,
e
S
0
)) = P [
e
S
p
( f
′
,
e
S
0
)]
if f is equivalent f
′
. According statement 2 all equiv-
alence classes are of the same size. Let n
c
be number
of permutations in each equivalence class. Then
|
e
S
T
(P
pv
)| ∗ n
c
|
e
S
b
| ∗ n
c
=
=
| f ∈
e
f|Q
min
(
e
S
p
( f, F),
e
M
c
) < Q
min
(
e
S
0
,
e
M
c
)}|
|
e
f|
.
Q.E.D.
Thus ratio
|{ f ∈
e
f|Q
min
[
e
S
p
( f, F),
e
M
c
] < Q
min
(
e
S
0
,
e
M
c
)}|
|
e
f|
(3)
theoretically allows to calculate exact p-value testing
validity of null hypothesis about exhaustive descrip-
tion of existing regularity by simple regularity from
e
M
s
. But practically it is impossible to calculate exact
p-values because of huge amount of possible permu-
tation. However it is easily to estimate 3 using rela-
tively small number of random permutations that are
generated by random numbers generator. Let
e
f
g
= { f
j
| j = 1, . . . , N
g
}
be set of permutations calculated by by random num-
bers generator. Then p-value may be estimated as ra-
tio
|{ f
j
∈
e
f
g
|Q
min
(
e
S
p
( f, F),
e
M
c
) < Q
min
(
e
S
0
,
e
M
c
)}|
N
g
(4)
2.3 Choice of Simple Model
Technique described in previous subsection may
be used only if simple model from
e
M
s
has been
previously chosen. Supposition 1 declares that com-
plicated model must not be used when there is simple
model that exhaustively describes data. Such model
may be searched by evaluating all predicting func-
tions from
e
M
s
with the help of described in previous
section PT version. But it is practically impossible
to implement such approach. In this paper only two
simple predicting functions from
e
M
s
are evaluated.
At first simple predicting function is studied that
is searched with the help of standard least squares
technique. It is naturally to hope that in many task LS
regression is very close to a model that exhaustively
describes data. However experiments with optimal
valid partitioning method (Senko and Kuznetsova,
2006) demonstrated that really false complicated
regularity R
c
may be mistakenly evaluated as valid.
Such mistakes take place when regularity are verified
relatively simple regularity R
s
that in the best way
approximate data. But at that R
s
significantly deviates
from verified complicated regularity R
c
. So a method
was developed in (Kuznetsova et al., 2013) verifying
more complicated model R
c
relatively simple model
that minimally deviates from R
s
.
Let try to explain why such technique may be
useful. Suppose that F
s
(x) is some predicting func-
tion from
e
M
s
, F
o
c
(x) is argmin
F(x)∈
e
M
c
Q[
e
S
0
, F(x)],
AMethodforEvaluatingValidityofPiecewise-linearModels
439

δ( j) = F
s
(x
j
) − F
o
c
(x
j
).
Discussed approach is based on evaluating upper
boundary of Q
min
[
e
S
p
( f, F
s
),
e
M
c
] where f ∈
e
f.
But by definition of
e
S
p
( f, F
s
)
Q
min
[
e
S
p
( f, F
s
),
e
M
c
] < Q[
e
S
p
( f, F
s
), F
o
c
] =
=
m
∑
j=1
[r
f( j)
+ F
s
(x
j
) − F
o
c
(x
j
)]
2
=
m
∑
j=1
[r
f( j)
+ δ( j)]
2
=
=
m
∑
j=1
r
2
f( j)
+ 2
m
∑
j=1
δ( j)r
f( j)
+
m
∑
j=1
δ
2
( j).
On another hand
Q
min
(
e
S
o
,
e
M
c
) = Q(
e
S
o
, F
o
c
) =
=
m
∑
j=1
[y
j
− F
o
c
(x
j
)]
2
=
m
∑
j=1
[y
j
− F
s
(x
j
)+
+F
s
(x
j
) − F
o
c
(x
j
)]
2
=
m
∑
j=1
[r
j
+ δ( j)]
2
=
m
∑
j=1
r
2
j
+ 2
m
∑
j=1
δ( j)r
j
+
m
∑
j=1
δ
2
( j).
Taking into account that
m
∑
j=1
r
2
j
=
m
∑
j=1
r
2
f( j)
we receive that
Q[
e
S
p
( f, F
s
), F
o
c
] − Q
min
(
e
S
o
,
e
M
c
) =
= 2
m
∑
j=1
δ( j)[r
f( j)
) − r
j
] ≤ 2
m
∑
j=1
|δ( j)| · |[r
f( j)
) − r
j
]|.
Thus upper bound for Q
min
[
e
S
p
( f, F
s
),
e
M
c
] tends to
Q
min
[
e
S
o
,
e
M
c
] as max
j=1,...,m
|δ
j
| tends to 0. It is more
probable that inequality
Q
min
[
e
S
p
( f, F
s
),
e
M
c
] < Q
min
[
e
S
o
,
e
M
c
]
is true when max
j=1,...,m
|δ
j
| is small. So we may
hope that p-value that is calculated by ratios 4 will
be greater when max
j=1,...,m
|δ
j
| is small. Thus small
p-value received when F
c
o
is verified relatively clos-
est simple model is strong argument for absence of
simple model from
e
M
s
that cannot be rejected using
complicated model. Existence of such argument cor-
responds to Supposition 1 correctness.
3 APPLICATION EXAMPLE
3.1 Objectives
Effect of vitamin D status(vitD) on parathyroid hor-
mone (PTH) concentration was studied (Kim et al.,
2012). Now serum 25 (OH) D is the best indica-
tor of the (vitD), but target levels of vitamin D in
the blood are still represent a matter of debate. So
the priority arrears of the research are the develop-
ment of a method-dependent reference values with
the use of biomarkers for vitD sufficiency. One such
widely recognized biomarker is the correlation of
vitD with PTH. But supposition exists that vitD cor-
relates with PTH only when vitD concentration is less
than certain threshold level and there is correlation
”loss” when vitD concentration is higher than thresh-
old level. Goal of our research was statistical verifica-
tion of last supposition and search of optimal model
that describes dependence of PTH on vitD. It must be
noted that discussed supposition corresponds to use
of piecewise-linear model.
3.2 Data Set
The study included patients (n = 139, males 18%,
mean age 48,5 ± 18 years) in which levels of to-
tal 25(OH)D (LIAISON, DiaSorin) and PTH (ELEC-
SYS, Roche) were measured during autumn period
(September-October). In selection of patients we
used exclusion criteria: presence of primary hyper-
parathyroidism, secondary or tertiary hyperparathy-
roidism on the backgroundterminal chronic renal fail-
ure, blood creatinine level of more than 100 mmol/l
or GFR less than 60 ml/min/1,73m2, intake of active
vitD metabolites within 1 month prior the blood test.
3.3 Search of Optimal Regression
It is supposed that response variable Y is predicted
by variable X with the help of piecewise-linear model
with 2 segments
Y = β
l
0
+ β
l
1
X + ε
l
, whenX ≤ B
Y = β
r
0
+ β
r
1
X + ε
r
, whenX ≥ B (5)
At that it is supposed that
β
l
0
+ β
l
1
B = β
r
0
+ β
r
1
B. (6)
Let
e
M
B
pwl
be family of all piecewise-linear predicting
functions with 2 segments and fixed B . For each B re-
gression coefficients β
l
0
, β
l
1
, β
r
0
, β
r
1
are calculated from
observations
(y
1
, x
1
), . . . , (y
m
, x
m
)
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
440
with the help of standard least squares technique. It is
evident that search of coefficients may be reduced to
of quadratic programming task:
Q(
e
S
0
,
e
M
B
pwl
) =
∑
x
j
<B
(y
j
− β
l
0
− β
l
1
x
j
)
2
+
+
∑
x
j
<B
(y
j
− β
r
0
− β
r
1
x
j
)
2
→ min, (7)
when constraint (6) is satisfied. Partial derivatives of
Lagrange function
Q(
e
S
0
,
e
M
B
pwl
) + λ(β
l
0
+ β
l
1
B− β
r
0
− β
r
1
B)
by coefficients β
l
0
, β
l
1
, β
r
0
, β
r
1
must be equal 0 for the
task (7). Let
e
X
m
= {x
1
, . . . , x
m
}. Using 4 equalities
for partial derivatives and constraint (6) we receive
system of 5 equations.
m
l
β
l
0
+
¯
X
l
β
l
1
−
1
2
λ =
¯
Y
l
¯
X
l
β
l
0
+ d
l
x
β
l
1
−
B
2
λ = c
l
xy
m
r
β
r
0
+ d
r
x
β
r
1
+
1
2
λ =
¯
Y
r
¯
X
r
β
r
0
+ d
r
x
β
r
1
+
B
2
λ = c
r
xy
−β
l
0
− β
l
1
B+ β
r
0
+ Bβ
r
1
= 0
(8)
where
• m
l
is number of points in
e
X
m
satisfying inequality
x
j
< B,
• m
r
is number of points in
e
X
m
satisfying inequality
x
j
> B,
• d
l
x
=
∑
x
j
<B
x
j
,
¯
X
r
=
∑
x
j
>B
x
j
,
• c
l
xy
=
∑
x
j
<B
x
j
y
j
, c
r
xy
=
∑
x
j
>B
x
j
y
j
,
•
¯
X
l
=
∑
x
j
<B
(x
j
)
2
,
¯
X
r
=
∑
x
j
>B
(x
j
)
2
,
Optimal regression coefficients belongs to solution of
system (8). Let
e
X
c
= {x
c
j
′
j
′′
=
x
j
′
+ x
j
′′
2
|x
j
′
6= x
j
′′
, x
j
′
∈
e
X
m
, x
j
′′
∈
e
X
m
}
be a set of boundaries separating neighbour points
from
e
X
m
. To find LS piecewise-linear regression it
is sufficient to calculate Q(
e
S
0
,
e
M
B
pwl
) for all boundary
points from
e
X
c
and to select boundary corresponding
to minimal Q(
e
S
0
,
e
M
B
pwl
).
3.4 Data Analysis Results
Let x
vd
be concentration of serum 25(OH)D, y
ph
be concentration of PTH, y
lph
= logy
ph
. Optimal
piecewise-linear regressions calculating y
ph
and y
lph
were chosen in
e
M
pwl
with the help of technique de-
scribed in previous section. Optimal boundary point B
was equal 23.95
ng
ml
for model predicting y
ph
from x
vd
(task I) and B = 24.7
ng
ml
for piecewise-linear regres-
sion predicting y
lph
from x
vd
(task II). Dependence of
Q(
e
S
0
,
e
M
B
pwl
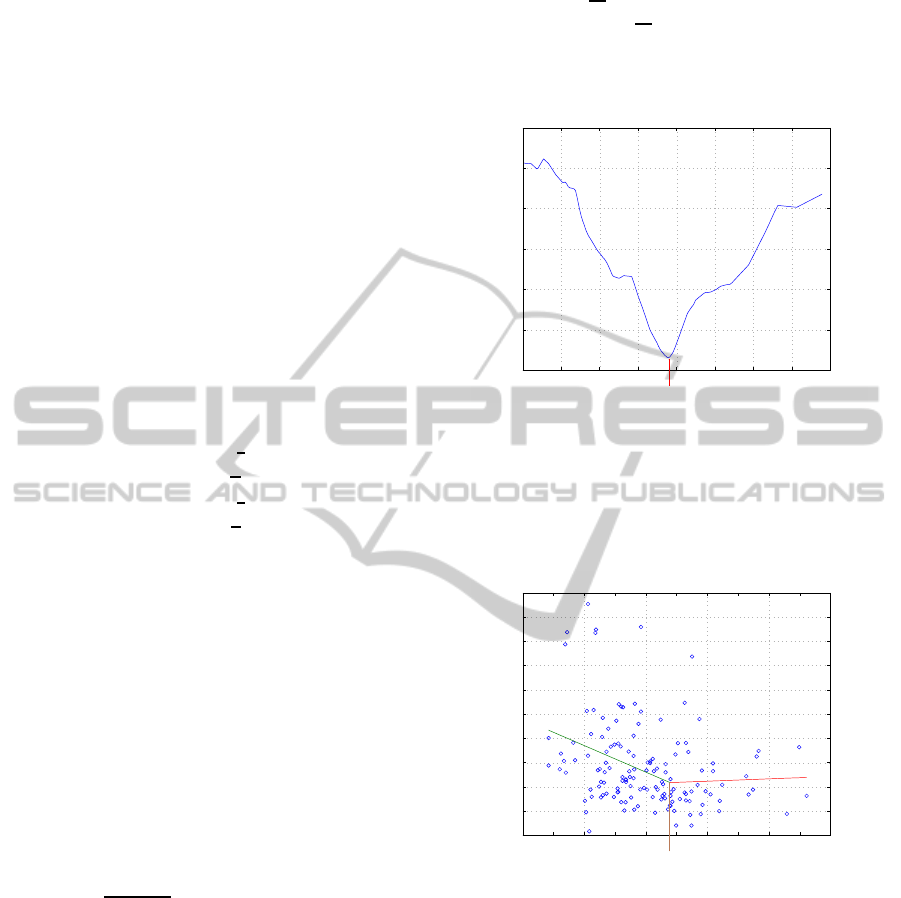
) on B in task I is given at figure 1.
5 10 15 20 25 30 35 40 45
1.51
1.52
1.53
1.54
1.55
1.56
1.57
B (ng/mL)
Q(B)
23.95
Figure 1: Dependence Q(
e
S
0
,
e
M
B
pwl
) on B in task I.
It is seen from figure 1 that point 23.95(ng/ml)
corresponds unique expressed global minimum of
Q(
e
S
0
,
e
M
B
pwl
). Graphic of piecewise-linear function
from model I is represented at figure 2. It is seen
0 5 10 15 20 25 30 35 40 45 50
0
20
40
60
80
100
120
140
160
180
200
25(OH)D (ng/mL)
PTH (pg/mL)
23.95
Figure 2: Optimal piecewise regression for task I.
that slope of linear predicting function inside left seg-
ment significantly exceeds slope of linear predicting
function inside right segment. Correlation coefficient
between y
ph
and x
vd
in group of patients with x
vd
<
23.95 is equal -0.2934 (significant at p¡0.01). Corre-
lation coefficient in group of patients with x
vd
> 23.95
is close to zero (equal 0.0351). Such results are in
good agreement with supposition that vitD correlates
with PTH only when vitD concentration is less than
certain threshold level. However statistical signifi-
cance of such correlation analysis is not too high be-
cause correlation coefficients are calculated in groups
formed by boundary B that was previously found by
the same data set. Let try to validate result with the
AMethodforEvaluatingValidityofPiecewise-linearModels
441

help of procedures verifying complicated models rel-
atively simple models that were discussed in previous
sections.
3.5 Verification
At the first stage null hypothesis about independence
of y
ph
on x
vd
was tested with the help previously dis-
cussed in (Senko and Kuznetsova, 2006) permutation
test version. Set of random permutations of integers
1, . . . , m was formed with the help of random num-
bers generator. This set
e
f
rng
consisted of N
g
elements.
Data sets {
e
S
p
( f
j
)| f
j
∈
e
f
rng
} was built from
e
S
0
by ran-
dom permutation of y
ph
positions relatively fixed po-
sitions of x
vd
. Statistical validity of null hypothesis is
evaluated with the help of p-value that is equal ratio
|{ f
j
∈
e
f
rng
|Q
min
[
e
S
p
( f
j
),
e
M
pwl
] < Q
min
(
e
S
0
,
e
M
pwl
)}|
N
g
.
In other words p-value is calculated as fraction of
random data sets where dependence of y
ph
on x
vd
is approximated better than at initial set
e
S
0
. Values
Q
min
(
e
S
0
,
e
M
pwl
) and Q
min
[
e
S
p
( f
j
),
e
M
pwl
] are calculated
with the help of procedure that is describe in section
3.3. Piecewise-linear modeling of y
ph
from x
vd
allows
to reject null hypothesis with p-value equal 0.000041.
Piecewise-linear modeling of y
lph
from x
vd
allows to
reject null hypothesis with p-value equal 0.000079.
At that number of random permutations was equal
10
6
. Then piecewise-linear regressions were veri-
fied relatively simple regression models. Optimal
piecewise-linear regression y
ph
= F
o
pwl
(x
vd
)+ε
pw
was
verified by testing null hypothesis about exhaustive
description of dependence by simple linear regression
y
ph
= α
0
+ α
1
x
vd
+ ε
1
. Piecewise-linear regression
y
lph
= F
o
pwl
x
vd
+ ε
pw
was verified by testing null hy-
pothesis about exhaustive description of dependence
by simple linear regression y
lph
= α
l
0
+ α
l
1
x
vd
+ ε
2
.
Two ways of regression coefficients α
0
, α
l
0
, α
1
, α
l
1
calculating were considered:
• simple regression coefficients were searched with
the help of standard LS procedure,
• such simple regression coefficients were chosen
that distance between verified piecewise-linear re-
gression and simple regression was minimal.
Let suppose that x values in
e
S
0
belong to some in-
terval (a
l
, a
h
). Then distance between two predicting
functions F
1
(x) and F
2
(x) is calculated by formula
D[F
1
(x), F
2
(x)] =
Z
a
h
a
l
[F
1
(x) − F
2
(x)]
2
dx.
Ratio (4) was used to estimate p-values. At that num-
ber of permutations was equal 10
6
. Results of verifi-
cation are represented in table.
Table 1: Results of verification.
target type of symple model p-value
y
ph
standard LS 0.022
y
lph
standard LS 0.026
y
ph
most close to F
o
pw
0.015
y
lph
most close to F
o
pw
0.0218
It is seen from table that p-values for null hypothe-
ses about exhaustive description of data by simple re-
gressions do not exceed 0.026. This result is strong
argument that simple regressions are not sufficient to
explain data and more complicated piecewise-linear
regression models are really necessary. Thus suppo-
sition that vitD correlates with PTH only when vitD
concentration is less than certain threshold level is sta-
tistically valid.
4 CONCLUSIONS
So method was proposed that allows to evaluate valid-
ity of choice between simple or complicated regres-
sion models in terms of p-values. Method is based on
testing null hypothesis about independence of devia-
tions from simple predicting function on X variables.
Method was successfully used for evaluating correct-
ness of biomedical supposition that vitamin D status
correlates with parathyroid hormone levels. Method
may be used in variety of tasks where a problem of
choice between more complicated or simple models.
REFERENCES
Akaike, H. (1974). A new look at the statistical model iden-
tification. IEEE Transactions on Automatic Control,
vol.19, iss.6,:pp. 716–723.
Domingos, P. (1999). The role of occam’s razor in knowl-
edge discovery. Data Mining and Knowledge Discov-
ery, vol. 3, iss. 4:pp. 409–425.
Ernst, M. (2004). Permutation methods: A basis for exact
inference. Statistical Science, 19(4):676–685.
Golland, P. et al. (2000). Permutation test for classification.
Journal of Machine Learning Research, 1.
Good, P. (2005). Permutation, Parametric and Bootstrap
Tests of Hypotheses. Springer Science+Business Me-
dia, Inc.
Hannan, E. and Quinn, B. (1979). The determination of the
order of an autoregression. Journal of the Royal Sta-
tistical Society, Series B (Methodological), vol.41:pp.
190–195.
Kim, G. et al. (2012). Relationship between vitamin d,
parathyroid hormone, and bone mineral density in el-
derly koreans. J Korean Med Sci.
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
442

Kim, H.-J. et al. (2000). Permutation tests for joinpoint
regression with applications to cancer rates. Statist.
Medicine, vol.19.
Kuznetsova, A. et al. (2013). Modification of the method
of optimal valid partitioning for comparison of pat-
terns related to the occurrence of ischemic stroke in
two groups of patients. Pattern Recognition and Im-
age Analysis, 22(4):10–25.
Ojala, M. and Garriga, G. (2010). Permutation tests for
studying classifier performance. Journal of Machine
Learning Research.
Rissanen, J. (1978). Modeling by shortest data description.
Automatica, vol. 14, iss. 5:pp. 465–658.
Schwarz, G. (1978). Estimating the dimension of a model.
Annals of Statistics, vol. 6:pp. 461–464.
Senko, O. and Kuznetsova, A. (2006). The optimal valid
partitioning procedures. InterStat, Statistics in Inter-
net,http://ip.statjournals.net.
AMethodforEvaluatingValidityofPiecewise-linearModels
443
