
Model-based Approach to Tissue Characterization using Optical
Coherence Tomography
Cecília Lantos
1,2,3
, Rafik Borji
1
, Stéphane Douady
2
, Karolos Grigoriadis
1
,
Kirill Larin
4
and Matthew A. Franchek
1
1
Department of Mechanical Engineering, University of Houston, 4800 Calhoun Road, Houston, TX 77004, U.S.A.
2
Laboratory of Matter and Complex Systems, Paris Diderot University, 5 Rue Thomas Mann, Paris, 75013, France
3
Department of Hydrodynamic Systems, Budapest University of Technology and Economics,
Műegyetem Rakpart 3-9, Budapest, 1111, Hungary
4
Department of Biomedical Engineering, University of Houston, 4800 Calhoun Road, Houston, TX 77004, U.S.A.
Keywords: Optical Coherence Tomography, Tissue Characterization, Medical Diagnostics, Cancer, Liposarcoma,
Model-based, Imaging.
Abstract: Structural property of the tissue can be quantified by its optical scattering properties. Since a tumor is
differentiated from healthy tissue based on morphological analysis, model-based approach to cancer
diagnosis is developed. The scattering property is measured using Optical Coherence Tomography. The
structural subsurface images from the measurements are described quantitatively. A parametric model is
developed to classify tissue as healthy or cancerous. A statistical model-based imaging method is created to
distinguish healthy vs. cancerous soft tissue using the example of human Normal Fat vs. Well-
Differentiated- (WD-), and De-Differentiated Liposarcoma (DDLS).
1 INTRODUCTION
Characteristics of tissue structural properties are
studied non-invasively with different imaging
modalities (Magnetic Resonance Imaging,
Computed Tomography, Ultrasound, Optical
Coherence Tomography …) (Rembielak, 2011;
Morris, 2012). Each works at different scale
depending on interest based on different physical
principles using specific frequency range of the
electromagnetic spectrum (radio frequency-, X-ray,
sound-, light wave…). These techniques can be
coupled for multidisciplinary analysis of the tissue
providing different information detected from
backscattered waves from the internal structure. The
outputs of these backscattered signals are grayscale
images with different resolution and imaging depth
revealing the subsurface structure (Rembielak, 2011;
Morris, 2012).
We chose Optical Coherence Tomography
(OCT) to analyze tissue structural properties. OCT
records images based on near infrared (NIR) laser
light scattered back from the tissue (Drexler, 2008,
Brezinski, 2006). Instead of subjective image
analysis, we approach the diagnosis from
mathematical point of view in order to quantify
topological changes. We develop a simple statistical
model based on the images analyzing the scattering
properties distinguishing various tissue types. The
tissue example is Normal Fat tissue vs. Well-
differentiated and De-differentiated Liposarcoma,
but the idea can be broadened toward the analysis of
other type of cancer since the diagnosis is based on
morphology. This model based imaging can become
a clinical tool to provide a second opinion for
physiologists.
In the literature, some approaches have been
elaborated that could differentiate quantitatively
between the various tissue types and specifically
between healthy and cancerous tissue recorded with
OCT. The attenuation of backscattered laser light in
function of depth (z) in the biological material
theoretically follows an exponential function
(Drexler, 2008; Brezinski, 2006):
I
z
I
e
(1)
defined by the scattering coefficient
characterizing different tissue types, calculating
from the slope of the intensity attenuation in dB
scale. This implies the abstraction of the tissue
19
Lantos C., Borji R., Douady S., Grigoriadis K., Larin K. and A. Franchek M..
Model-based Approach to Tissue Characterization using Optical Coherence Tomography.
DOI: 10.5220/0004805500190027
In Proceedings of the International Conference on Bioimaging (BIOIMAGING-2014), pages 19-27
ISBN: 978-989-758-014-7
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

structure. For inhomogeneous material the slope is
calculated by averaging or filtering, and the analysis
of the deviation from the slope characterizes well the
tissue (Lev, 2011, Yang, 2011, McLaughlin, 2010,
Mujat, 2009, Goldberg, 2008).
Morphological pattern of breast cancer in OCT
images has been studied by fractal analysis using
box-counting algorithm (Sullivan, 2011). The
periodicity analysis based on the scattering effect
due at the cell boundaries distinguishes healthy vs.
cancerous breast tissue (Zysk, 2006; Mujat, 2009;
Goldberg, 2008). A similar method based on cell
counting is already applied on OCT images of
Liposarcoma (Carbajal, 2011).
Speckle phenomena are a random scattering
effect, called texture, which analysis reveals the
tissue types in case of inadequate structural
resolution (Gossage, 2003; Gossage, 2006).
2 TISSUE STRUCTURE
RECORDED WITH OPTICAL
COHERENCE TOMOGRAPHY
Optical Coherence Tomography (OCT) is a well-
known structural imaging method applied on
biological material, in particular for cancer diagnosis
(Drexler, 2008, Brezinski, 2006). OCT has a better
resolution (3-10 μm) compared to other diagnostic
methods, revealing the subsurface structure in a 1-3
mm deep region under the surface, and has proved to
be the most suitable imaging method applied on
Liposarcoma (Carbajal, 2011; Lahat, 2009; Lev,
2011).
According to the WHO report on Soft tissue
tumors, Liposarcoma is part of the Adipocytic
Tumors. In this study we differentiate Normal Fat
from Intermediate (locally agressive) tumor, so
called Well-Differentiated Liposarcoma (WDLS)
and from one type of Malignant tumor (having risk
to metastasize), called De-Differentiated
Liposarcoma (DDLS) (Fletcher, 2006).
Tissue samples were excised from human
patients’ abdomen/retroperitoneum at the University
of Texas M. D. Anderson Cancer Center
(UTMDACC). Protocols for tissue processing were
approved by the UTMDACC and University of
Houston Biosafety Committees. Histological
diagnosis and classification of samples was
performed by a UTMDACC sarcoma pathologist.
The tissue was put in sterile phosphate buffered
saline then stored in refrigerator until imaged with
the OCT system.
We record the tissue on a Spectral-Domain (SD)
OCT measuring rig in the BioOptics Laboratory at
the University of Houston. A supraluminescent laser
diode (Superlum, S840-B-I-20) generates a
broadband laser signal with center wavelength at λ
0
= 840 nm, spectral bandwidth at Δλ = 50 nm and
output power at 20 mW (Carbajal, 2011) (Figure1).
Figure 1: a) Normal Fat Tissue cross-section OCT image
(mature fat, adipose cells), logarithmic response. b) Well-
differentiated Fat Tissue cross-section OCT image (WDLS
with extensive myxoid change), logarithmic response. c)
De-differentiated Fat Tissue cross-section OCT image
(Highly fibrotic DDLS), logarithmic response.
The above images show the cross-section of
Human Normal Fat tissue (Figure 1a), WDLS
(Figure 1b) and DDLS (Figure 1c). This 2D cross-
section called B-scan is composed of 500 adjacent
A-lines. One A-line (1D) shows the backscattered
intensity variation in function of depth from a laser
footprint of 8 μm in focal plane. The region is 3mm
BIOIMAGING2014-InternationalConferenceonBioimaging
20
wide scanned with a galvanometer mirror, with
backscattered light collected from a region of up to
~1 mm in depth.
The internal structure is revealed. We can see the
differences of the different tissue types on the gray-
scale images. We intend to transform the qualitative
information from the images to a quantitative
statistical parametric description of the tissue. The
statistical model is based on the variability of the A-
lines in the cross-section at a given region. One A-
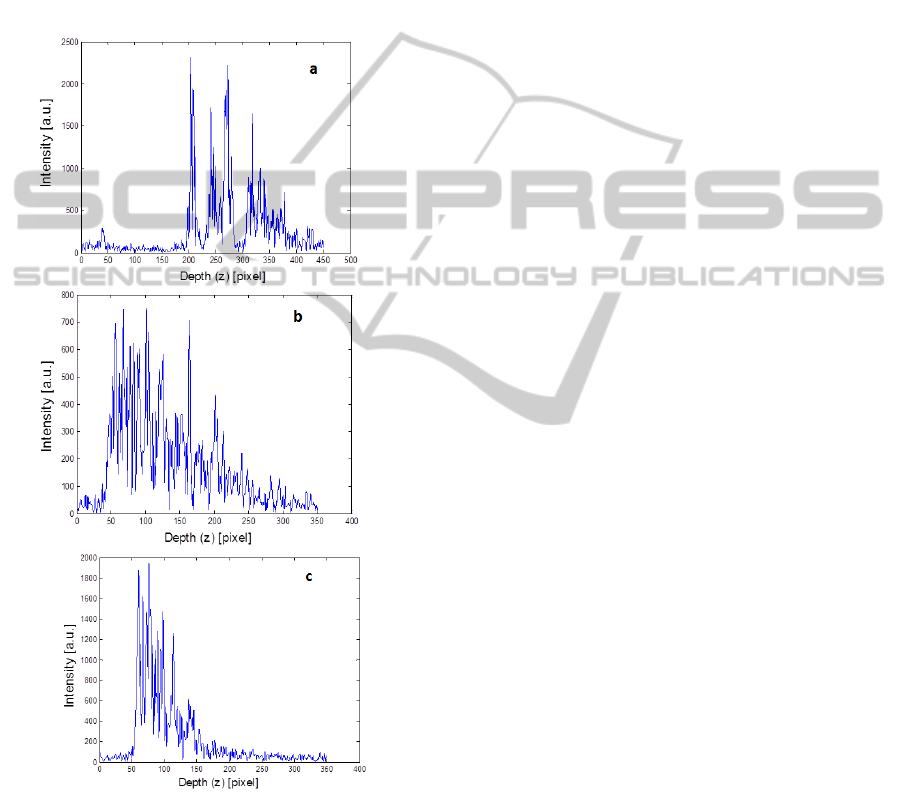
line of the different tissue types is seen on Figure 2.
Figure 2: OCT A-line of a) Normal Fat b) WDLS c)
DDLS. The intensity of the input laser light and the path-
length difference between the reference mirror and the
tissue surface differ in each case.
1D imaging (Intensity as a function of depth at a
single point) is obtained by applying Digital Signal
Processing methods on the data detected on a line
scan camera (Basler Sprint L104K-2k, 2048 pixel
resolution, 29.2 kHz line rate), -with a resolution of
2k, and a 10x10 μm2 pixel size, detecting 2048
wavelength intensity values between 800-890 nm.
The signal is digitized using an analog-to-digital
converter (NI-IMAQ PCI-1428). The intensity
detected on the line-scan camera is the cross-
correlation of the broadband laser light electric field
split in a Michelson interferometer and scattered
back from a reference mirror and from the sample
layers at each frequency component (Carbajal, 2011,
Drexler, 2008, Brezinski, 2006). The broadband
laser light is decomposed into its spectral
components in passing through a diffraction grating
(Wasatch Photonics, 1200 grooves/mm). The
measurement setting and DSP is computed in
Labview, whence the intensity functions as function
of depth are analyzed in Matlab.
3 DATA ANALYSIS
The post-processing steps to determine the model on
the Fourier-domain signatures derived from OCT
data will be explained here. Human Normal Fat,
WDLS and DDLS tissue samples will be analyzed.
We will focus on the statistical properties of the
backscattered intensity signals.
For the computation, the tissue surface should
first be numerically straightened. We apply the
canny edge detector implemented in Matlab Image
Processing Toolbox on the B-scans after median
filtering the images. This can be used to align the
scans, but does not yield the absolute position of the
surface with respect to common origin. Before
further analysis we screen all the B-scans to verify
that each one is straightened properly.
At each depth position the mean and standard
deviation of the intensity signals will be calculated.
Then, attenuation effects are removed from the data
by dividing the Intensity Values or the standard
deviation of the A-lines by the mean from each
backscattered intensity response, so as to normalize
every scan line.
The images are corrected according to the
normalized camera sensitivity curve, to eliminate the
errors coming from the intensity variations because
of the oblique tissue surface. It is due to the camera
feature recording the same sample point at lower
intensity from farther path-length differences
following a Gaussian decay (Bajraszewski, 2008).
3.1 Standard Deviation over Mean
In the first case the tissue characterization will be
defined from the Probability Density Functions
Model-basedApproachtoTissueCharacterizationusingOpticalCoherenceTomography
21
(PDF) of the STD/MEAN curves. The three-
parameter Generalized Extreme Value (GEV)
Distribution fits the histograms well due to its high
flexibility:
y
f
x|k,μ,σ
exp
1k
1k
(2)
where x is the std/mean of the intemsity values, y is
the distribution, k is the shape-, σ is the scale-, μ is
the location parameter.
To find the tissue surface on the straightened
images the mean of the A-lines in one B-scan, and
the first derivatives of the mean are calculated from
the uncorrected images. The tissue surface is defined
at the highest derivative point.
After the tissue surface is defined, 150 pixel =
0.659 mm is considered for analysis because the
most dense tissue (DDLS) does not reflect light from
deeper region at this wavelength range and camera
settings (Figures 3a).
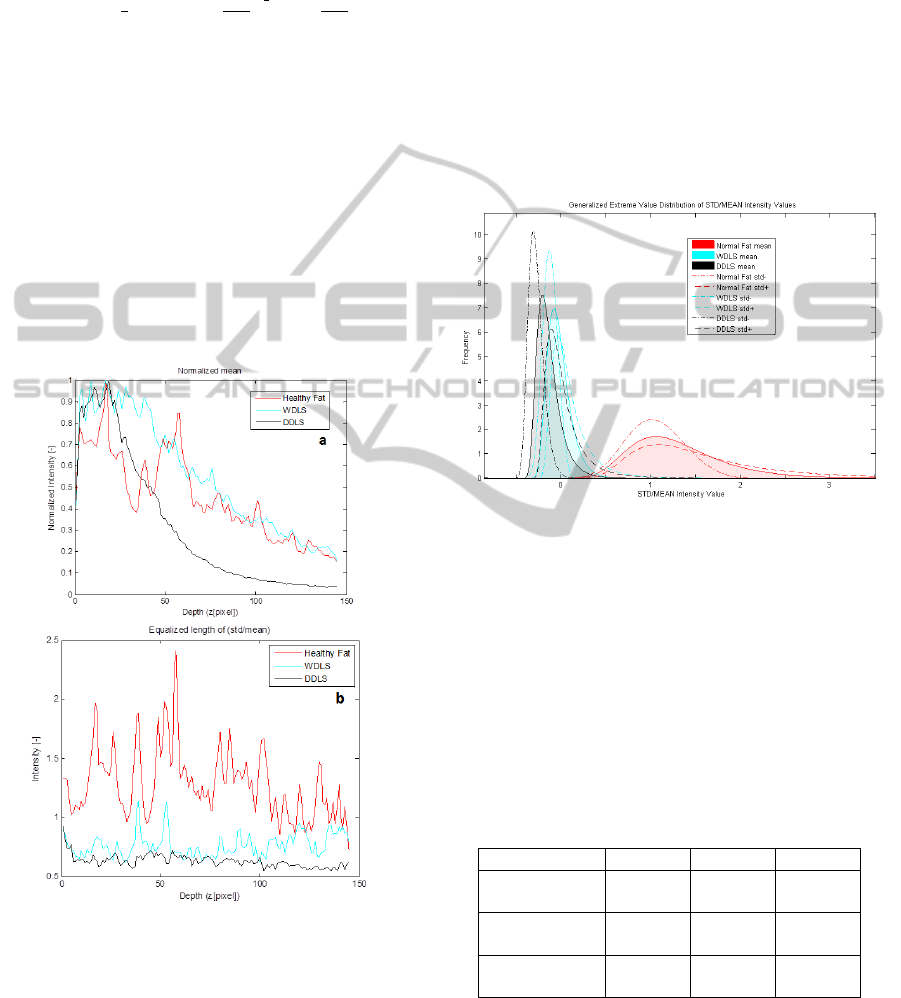
Figure 3: a) Averaged B-scan, mean intensity value at
each depth position on Normal Fat, WDLS, DDLS; (150
pixels from the tissue surface) The curves here are
normalized according to maximum value only for
representation. b) Standard Deviation over Mean at each
depth position in the same region.
The next step will be to define the Region of Interest
(ROI) on the curves for analysis. This analysis relies
on the use of a windowing scheme, in which
sections of the intensities as function of depth are
evaluated separately.
After evaluation of the data in each window
region at each B-scan via the parameters of the GEV
distribution, a window size of 40 pixel = 0.1758 mm
is chosen, beginning from the tissue surface. Our
method turned out to be independent on the surface
scattering effect.
To depict the accuracy of the results, 160 WDLS
or DDLS and 200 Normal Fat B-scans were
analyzed. Figure 4 shows the mean and STD of the
GEV parameters on the Gaussian corrected curves.
It characterizes well the different tissue types.
Figure 4: Histogram, GEV Distribution (k, σ, μ) calculated
from the STD/mean ratio of the intensity values at each
depth position in ROI, mean and standard deviation on
200 B-scans of Normal Fat, and 160 B-scans of WDLS
and DDLS.
The curve coefficients well differentiate between
the healthy and cancerous tissues, but there is less
distinction between the grades of the cancer (Table
1).
Table 1: GEV parameters calculated from the STD/MEAN
ratio of the intensity values at each depth position, mean
and standard deviation on 200 B-scans of Normal Fat, and
160 B-scans of WDLS and DDLS.
STD/MEAN k σ μ
Baseline
(Normal Fat)
0.0007
+0.2347
0.2151
+0.0579
1.2796
+0.0659
Deviation1
(WDLS)
-0.0128
+0.2443
0.0529
+0.0120
0.7093
+0.0359
Deviation2
(DDLS)
0.0857
+0.1673
0.0493
+0.0128
0.6502
+0.0584
To draw the deviation from the baseline tissue
the next parameters are calculated, where b is the
Baseline tissue parameter, d is the Deviated tissue
parameter.
BIOIMAGING2014-InternationalConferenceonBioimaging
22
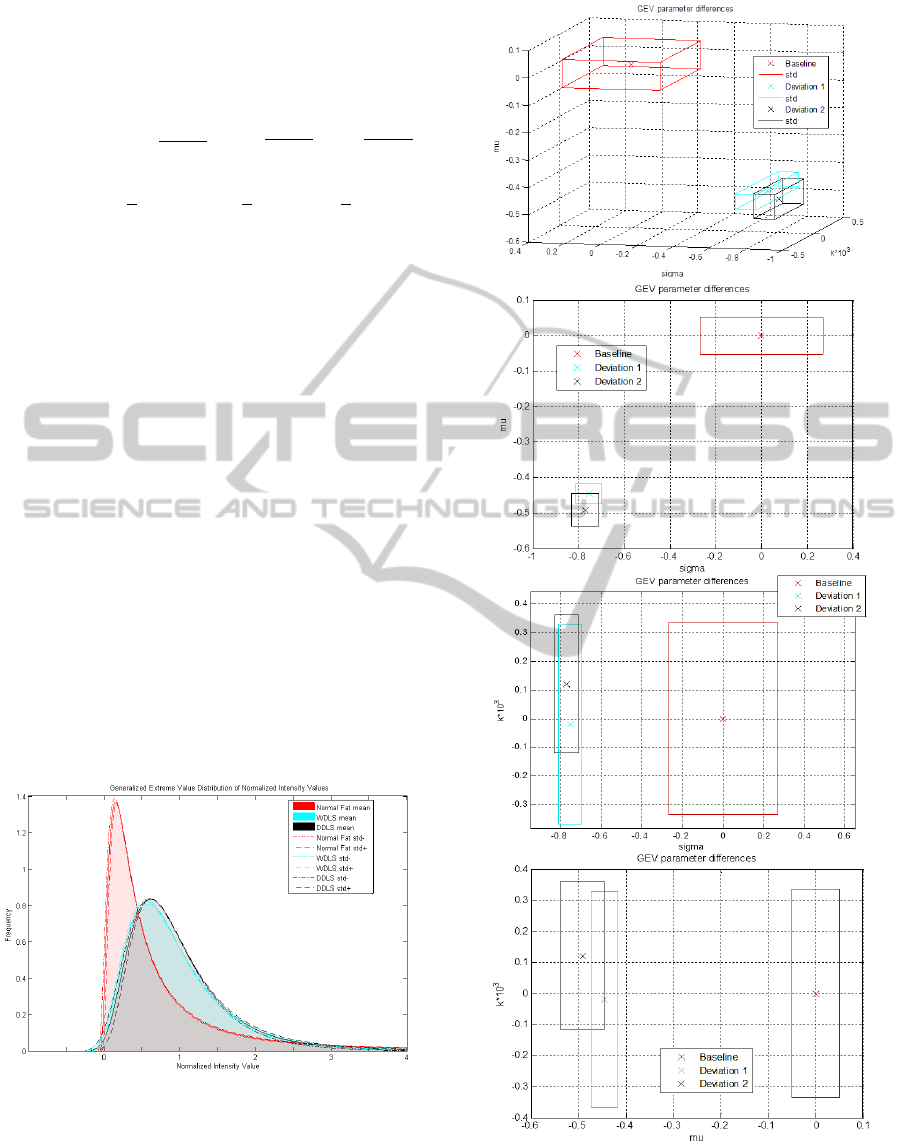
Table 2: Comparison of the GEV parameters calculated
from the STD/mean ratio of the intensity values at each
depth position, mean and standard deviation on 200 B-
scans of Baseline Tissue and 160 B-scans of Deviation
1&2.
STD/MEAN
∆
∆
∆
Baseline
(Normal Fat)
0
+335.2857
0
+0.2692
0
+0.0515
Deviation 1
(WDLS)
-19.2857
[-368.2857;
329.7143]
-0.7541
[-0.8099;
-0.6983]
-0.4457
[-0.4737;
-0.4176]
Deviation 2
(DDLS)
121.4286
[-117.5714;
360.4286]
-0.7708
[-0.8303;
-0.7113]
-0.4919
[-0.5375;
-0.4462]
The next figure represents the coefficient
differences on the axes of a 3D coordinate system. It
is clear, that there is a relevant separation between
the healthy and cancerous tissue in each projection
plane.
3.2 Normalized Intensity Variation
A second method is developed to analyze the same
data set. Instead of calculating the STD/MEAN, all
the measured intensity values are now considered,
and also normalized by the mean intensity at each
depth position. The same windowing process was
applied on the A-lines and B-scans, and the optimal
window size of 40 pixels beginning from the surface
has been proved. Figure 6 and Table 3 shows the
mean and STD of the GEV parameters
characterizing the different tissue types.
Figure 6: Histogram, GEV Distribution (k, σ, μ) calculated
from the mean-normalized intensity values in ROI, mean
and standard deviation on 200 B-scans of Normal Fat, and
160 B-scans of WDLS and DDLS.
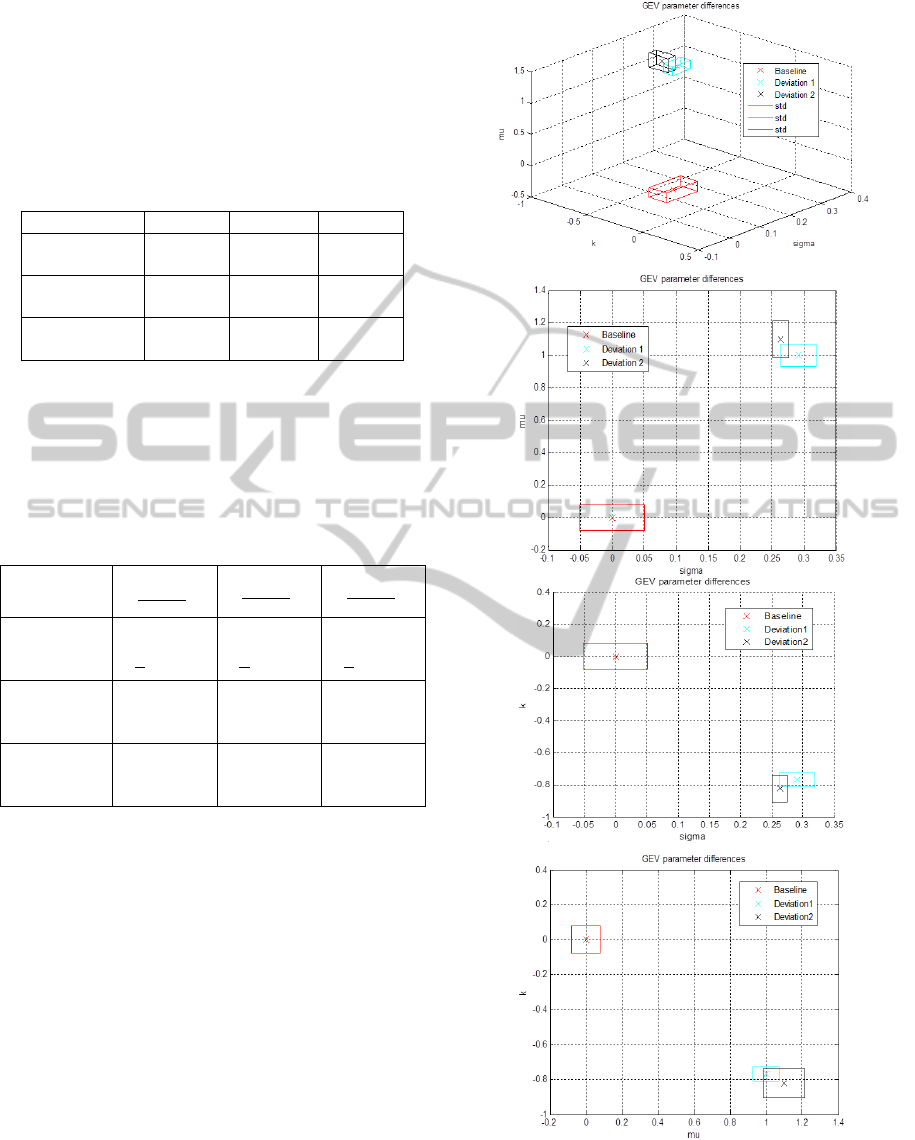
Figure 5: Comparison of the GEV parameters represented
at each axe of the 3D coordinate system calculated from
the STD/mean ratio of the intensity values at each depth
position, mean and standard deviation on 200 B-scans of
Baseline Tissue and 160 B-scans of Deviation 1&2.
Model-basedApproachtoTissueCharacterizationusingOpticalCoherenceTomography
23
Similarly to the first method the curve coefficients
characterize well the healthy and cancerous tissue
but WDLS and DDLS coefficients are not
sufficiently distinguished.
Table 3: GEV parameters calculated from the mean-
normalized intensity values in ROI, mean and standard
deviation on 200 B-scans of Normal Fat, and 160 B-scans
of WDLS and DDLS.
I/MEAN k σ μ
Baseline
(Normal Fat)
0.8209
+0.0647
0.3532
+0.0181
0.3191
+0.0254
Deviation1
(WDLS)
0.1905
+0.0358
0.4561
+0.0100
0.6381
+0.0229
Deviation2
(DDLS)
0.1447
+0.0684
0.4462
+0.0044
0.6700
+0.0364
The next table shows the deviation from the
baseline tissue, where b is the Baseline tissue
parameter, d is the Deviated tissue parameter.
Table 4: Comparison of the GEV parameters calculated
from the mean-normalized intensity values in ROI, mean
and standard deviation on 200 B-scans of Baseline Tissue
and 160 B-scans of Deviation 1&2.
ΣI /MEAN
∆
∆
∆
Baseline
(Normal
Fat)
0
+0.0788
0
+0.0512
0
+0.0796
Deviation1
(WDLS)
-0.7679
[-0.8115;
-0.7243]
0.2913
[0.2630;
0.3196]
0.9997
[0.9279;
1.0715]
Deviation2
(DDLS)
-0.8237
[-0.9071;
-0.7404]
0.2633
[0.2508;
0.2758]
1.0997
[0.9856;
1.2137]
The next figure shows similar results then the
first method representing the coefficient differences
in the 3D coordinate system and separating well the
healthy and cancerous tissue in each projection
plane, however the different cancer tissues are
overlapped.
4 DISCUSSION
We can deduce that both statistical analyses are a
viable method to differentiate tissue types with a
good accuracy. The method is independent on the
measurement settings as the results are normalized
by the mean of the intensity values at each depth
position, and errors due to path-length differences
are corrected. For comparison the data analysis was
also applied on the images without this correction
Figure 7: Comparison of the GEV parameters represented
at each axe of the 3D coordinate system calculated from
the mean-normalized intensity values in ROI, mean and
standard deviation on 200 B-scans of Baseline Tissue and
160 B-scans of Deviation 1&2.
BIOIMAGING2014-InternationalConferenceonBioimaging
24
revealing that only the shape parameter (k) is
affected in a non-negligible way in the case the
STD/MEAN ratio is calculated. This analysis is
more sensitive also to the way how we find the
surface of
the tissue since the data points from
which the histogram is drawn is deduced calculating
the STD/MEAN from each depth position (40
pixels), comparing to the second method where the
histogram is drawn from the data points contained in
all the Region of Interest (40x200 or 40x160 pixel
points). In case we want to get absolute parameters,
which describe tissue type, the correction is needed.
The method could be developed to distinguish
better the different grade of cancer implementing
with additional factors, e.g. the mean intensity
values at each depth position. The weak point of the
measurements is that setting the position of the
tissue under the laser light to get a visible subsurface
structure is controlled manually. The measurements
revealed that the focus position does not affect
significantly the quantitative results, but some
saturated intensity points can also affect the
statistics.
It has already been proved that this non-invasive
measurement technique shows good similarities with
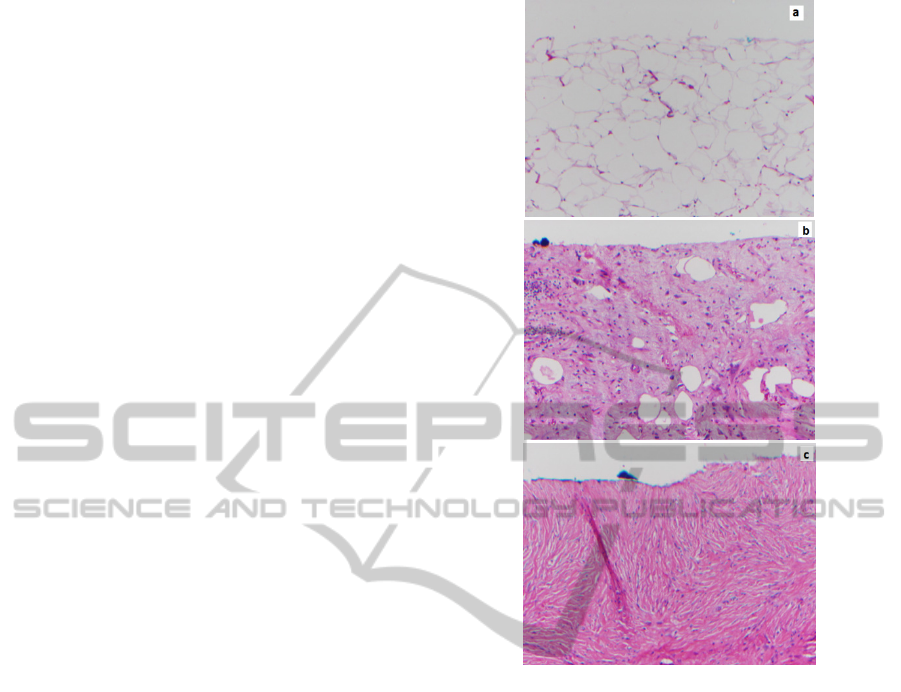
stained histology (Figure 8) (Lev, 2011). The
novelty of our study was to develop a mathematical
model-based approach instead of visual grading of
the structure to be able to differentiate tissue types.
The structure is detected from the scattering
properties of the tissue types. The laser clearly
reveals the adipose cells seen in Normal Fat. WDLS
has extensive myxoid change including vasculature,
but still has some adipose cells with varying size,
which is a diagnostic of WDLS. The part of DDLS
imaged here resembles fibrotic tissue.
Cancerous tissue is much denser than healthy
tissue. Since light scattering occurs chiefly at
interfaces, scattering is much stronger in cancerous
tissue. The inhomogeneous Normal Fat is
distinguished with periodic scattering at the cell
boundaries. The attenuation of light is higher in the
dense tissue, detectable with the attenuation
coefficient
, and the back reflection loses the
periodicity as the adipose cells dedifferentiate in the
cancerous tissue. The optical properties show the
morphology of the tissues, the scattering effects
reveal the cellular structure at a good resolution for
our analysis.
For medium grade sarcoma (WDLS), there is
much larger cell size dispersion than in healthy
tissue. An analysis of the structure’s periodicities is
sensitive to this, as well as speckle analysis. Cell
counting analysis can reveal the difference between
Figure 8: Histological images (Magnification 10x) of a)
Normal Fat b) Well-Differentiated Liposarcoma with
extensive mitotic change c) Highly Fibrotic De-
Differentiated Liposarcoma.
healthy tissue and high grade of cancer (DDLS), the
size of the variable adipose cells should be included
in the algorithm to distinguish between normal fat
and medium grade of cancer (WDLS). It can be
improved using artificial network analysis already
calculated on histological data (Sjöström, 1999).
There is a certain degree of order (on a given length
scale) in healthy tissue. On the contrary, in sarcoma,
many scales are present, which is revealed by fractal
analysis.
The techniques proposed in literature have not
been applied in clinical practice yet and there are
some shortcomings in the analysis. The slope
analysis discards all structural information; the
fractal analysis is too complicated and subject to
erroneous or ambiguous interpretation due to
experimental errors. The speckle analysis discards
information on loss of intensity due to multiple
scattering events. Finally it seems likely that
completely automated, practicable cell counting
Model-basedApproachtoTissueCharacterizationusingOpticalCoherenceTomography
25

analysis will not be achieved using traditional image
analysis software.
The aim of our study is to develop a simple
analysis technique based on a parametric method
that captures the structural features from the strength
of scattering. Here only one histological subtype of
WDLS and DDLS is described, however they can
represent several patterns (Miettinen, 2003,
Miettinen, 2010) The comparison of the different
histological subtypes and the ability to differentiate
from Normal Fat and Lipoma, benign adipose tissue
is a future study.
5 CONCLUSIONS
Our objective was to study the response of tissue to
a near infrared laser excitation, and, specifically, to
characterize differences between healthy and
cancerous tissue. The morphology of the subsurface
is depicted based on the backscattered near infrared
light. Parametric models of these backscattered
signal characteristics are derived and linked
statistically to the optical properties of Normal Fat,
Well-Differentiated Liposarcoma and De-
Differentiated Liposarcoma.
The accurate diagnosis at early stage of cancer,
as well as the recognition of the tumor boundary in
tissue is highly important. However OCT has been
well-recognized as a powerful method for cancer
detection from tissue morphology, the diagnosis
from these images is subjective and not obvious. We
intend to fill the need for an objective means of data
analysis. The goal of the current study was to
develop a quantitative diagnostic method
differentiating between healthy and cancerous tissue.
The data analysis is developed on images
recorded on human Normal Fat Tissue vs. Well-
differentiated (WD) and De-differentiated
Liposarcoma (DDLS). Further refinement will allow
to detect tumor boundary, diagnose other type of
cancer (e.g. breast cancer) where structural analysis
is required for diagnosis, or to monitor quantitatively
tumor progression during cancer therapy.
As a demonstration of these methods, statistical
analysis was developed to evaluate OCT images of
human fat specimens. An accurate result was found
to quantify healthy vs. cancerous tissue. The analysis
can be applied in real-time for diagnosis, and it is
much simpler comparing to other quantifying
method. This practical advantage gives a good
possibility to use in surgical evaluation.
We describe first time a model-based tissue-
characterization method based on structural
properties of healthy vs. cancerous tissue. Further
statistical validation, sensitivity/specificity analysis
and classification methods have to be performed on
other measurements to prove the efficacy of the
developed method.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the very important
contribution made by Shang Wang and Narendran
Sudheendran. This work was supported by the
Hungarian-American Fulbright Commission, the
French Ministry of Research and the University of
Houston. This work is the continuation of a poster
presentation held on November 16, 2012 at the
MEGA Research day of the University of Houston.
REFERENCES
Bajraszewski, T., Wojtkowski, M., Szkulmowski, M.,
Anna Szkulmowska, Huber, R., Kowalczyk, A., 2008.
Improved spectral optical coherence tomography using
optical frequency comb, Optics Express, 16(6).
Brezinski, M. E., 2006. Optical Coherence Tomography,
Principles and Applications, Academic Press Elsevier
Inc.
Drexler, W. and Fujimoto, J. G., 2008. Optical Coherence
Tomography: Technology and Applications, Springer,
Berlin, New York.
Carbajal, E. F., Baranov, S. A., Manne, V. G. R., Young,
E. D., Lazar, A. J., Lev, D. C., Pollock, R. E., Larin,
K.V., 2011. Revealing retroperitoneal liposarcoma
morphology using optical coherence tomography,
Journal of Biomedical Optics, 16(2).
Fletcher, C. D. M., Rydholm, A., Singer, S., Sundaram,
M., Coindre, J. M., 2006. Soft tissue tumours:
Epidemiology, clinical features, histopathological
typing and grading, WHO Classification of Soft Tissue
Tumors.
Goldberg, B. D., Iftimia, N. V., Bressner, J. E., Pitman, M.
B., Halpern, E., Bouma, B. E., Tearney, G. J., 2008.
Automated algorithm for differentiation of human
breast tissue using low coherence interferometry for
fine needle aspiration biopsy guidance, Journal of
Biomedical Optics, 13(1),
Gossage, K. W., Tkaczyk, T. T., Rodriguez, J. J., Barton,
J. K., 2003. Texture analysis of Optical Coherence
Tomography images: feasibility for tissue
classification, Journal of Biomedical Optics, 8(3).
Gossage, K. W., Smith, C. M., Kanter, E. M., Hariri, L. P.,
Stone, A. L., Rodriguez, J. J., Williams, S. K., Barton,
J. K., 2006. Texture analysis of speckle in Optical
Coherence Tomography images of tissue phantoms,
Physics in Medicine and Biology. 51.
BIOIMAGING2014-InternationalConferenceonBioimaging
26

Lahat, G., Madewell, J.E., Anaya, D. A., Qiao, W., Tuvin,
D., Benjamin, R. S., Lev, D. C., and Pollock, R. E.,
2009. Computed Tomography Scan-Driven Selection
of Treatment for Retroperitoneal Liposarcoma
Histologic Subtypes, Cancer 115(5).
Lev, D., Baranov, S. A., Carbajal, E. F. Young, E. D.,
Pollock, R. E., Larin, K. V., 2011. Differentiating
retroperitoneal liposarcoma tumors with optical
coherence tomography, Proceedings SPIE 7890,
Advanced Biomedical and Clinical Diagnostic
Systems IX, 78900U.
McLaughlin, R. A., Scolaro, L., 2010. Parametric imaging
of cancer with optical coherence tomography, Journal
of Biomedical Optics, 15(4).
Miettinen, M. M., 2003. Diagnostic Soft Tissue Pathology,
Churchill Livingstone.
Miettinen, M. M., 2010. Modern soft tissue pathology;
tumors and non-neoplastic conditions, Cambridge
University Press.
Morris, P., Perkins, A., 2012. Diagnostic imaging, Physics
and Medicine 2, Lancet, 379.
Mujat, M., R., Ferguson, D., Hammer, D. X., Gittins, C.,
Iftimia, N., 2009. Automated algorithm for breast
tissue differentiation in optical coherence tomography,
Journal of Biomedical Optics, 14(3).
Rembielak, A., Green, M., Saleem, A., 2011. Pat Price,
Diagnostic and therapeutic imaging in oncology,
Cancer Biology and Imaging, Medicine, 39(12).
Sjöström, P. J., Frydel, Wahlberg, L. U., 1999. Artificial
Neural Network-Aided Image Analysis System for
Cell Counting, Cytometry, 36.
Sullivan, A. C., Hunt, J. P., Oldenburg, A. L., 2011.
Fractal analysis for classification of breast carcinoma
in Optical Coherence Tomography, Journal of
Biomedical Optics, 16(6).
Yang, Y., Wang, T., Biswal, N.C., Wang, X., Sanders, M.,
Brewer, M., Zhu, Q., 2011. Optical scattering
coefficient estimated by optical coherence tomography
correlates with collagen content in ovarian tissue,
Journal of Biomedical Optics, 16(9).
Zysk, A. M., Boppart, S. A., 2006. Computational
methods for analysis of human breast tumor tissue in
optical coherence tomography images, Journal of
Biomedical Optics, 11(5).
Model-basedApproachtoTissueCharacterizationusingOpticalCoherenceTomography
27
