
Feature Selection by Rank Aggregation and Genetic Algorithms
Waad Bouaguel
1
, Afef Ben Brahim
1
and Mohamed Limam
1,2
1
LARODEC, ISGT, University of Tunis, Tunis, Tunisia
2
Dhofar University, Salalah, Oman
Keywords:
Rank Aggregation, Distance Function, Filter Methods, Feature Selection, Data Dimensionality.
Abstract:
Feature selection consists on selecting relevant features in order to focus the learning search. A simple and
efficient setting for feature selection is to rank the features with respect to their relevance. When several
rankers are applied to the same data set, their outputs are often different. Combining preference lists from
those individual rankers into a single better ranking is known as rank aggregation. In this study, we develop a
method to combine a set of ordered lists of feature based on an optimization function and genetic algorithm.
We compare the performance of the proposed approach to that of well-known methods. Experiments show that
our algorithm improves the prediction accuracy compared to single feature selection algorithms or traditional
rank aggregation techniques.
1 INTRODUCTION
The continued progress in developing different statis-
tical methods focusing on the same research question
have allowed investigating the search for ways to use
various methods simultaneously. To get the best re-
sult of all available alternatives, we need to integrate
their results in an efficient way. In the task of feature
selection, rank aggregation is an example of these in-
tegration methods.
Feature selection is an important stage of prepro-
cessing commonly used in data mining applications,
where a subset of the features that most contribute to
accuracy are selected. Feature selection is also a way
of avoiding the curse of dimensionality which occurs
when the number of availablefeatures signicantly out-
numbers the number of examples, as is the case in Bio
Informatics. Feature selection methods divide into
wrappers, filters, and embedded methods (Guyon and
Elisseff, 2003). Wrappers employ the learning algo-
rithm of interest as a black box to score subsets of
features according to their predictive power (Kohavi
and John, 1997). Filters select subsets of features as a
pre-processing step, independently of the chosen pre-
dictor. Embedded methods perform feature selection
in the process of training and are usually specic to
given learning algorithms. It is argued that, compared
to wrappers, filters are faster and that some filters pro-
vide a generic selection of features, not tuned for a
given learning algorithm. Another advantage of fil-
ters is that they can be used as a preprocessing step to
reduce space dimensionality and overcome overtting
(Guyon and Elisseff, 2003).
A particularly optimal implementation of filters
are methods that employ some criterion to score each
feature and provide a ranking (Caruana et al., 2003)
(Weston et al., 2003). From this ordering, several fea-
ture subsets can be chosen. This kind of the filter ap-
proach, that are known as rankers, can be extremely
efficient because they are quite simple. When apply-
ing multiple rankers based on different scoring cri-
teria we have often different feature ranking lists so
finding a way to aggregate them is required. Rank
aggregation is to combine ranking results of entities
from multiple ranking functions in order to generate a
better one (Borda, 1781) (Dwork et al., 2001).
Most of rank aggregation methods treat all the
ranking lists equally and give high ranks to those enti-
ties ranked high by most of the rankers. This assump-
tion may not hold in practice, however. For example,
for a given problem, a ranking list may give better
classification results than the others. It is not reason-
able to treat the results of all ranking lists equally.
To deal with the problem, weighted rank aggre-
gation techniques can be employed where different
rankers are assigned different weights. For example,
the weights can be calculated based on the mean av-
erage precision scores of the base rankers. In this
paper we investigate the use of ranking lists impor-
tance, based on their corresponding classification ac-
74
Bouaguel W., Ben Brahim A. and Limam M..
Feature Selection by Rank Aggregation and Genetic Algorithms.
DOI: 10.5220/0004518700740081
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval and the International Conference on Knowledge
Management and Information Sharing (KDIR-2013), pages 74-81
ISBN: 978-989-8565-75-4
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

curacy, in addition to feature weights in order to high-
light ranking lists that give better classification results
and features that are more relevant than others even
if they are equally ranked by different rankers. These
parameters are used by a genetic algorithm that re-
ceives multiple ranking lists and generate a final ro-
bust feature ranking list.
2 RANK AGGREGATION
Feature ranking consists on ranking the features with
respect to their relevance, one selects the top ranked
features, where the number of features to select is
specied by the user or analytically determined.
Many feature selection algorithms include feature
ranking as a principal selection mechanism because of
its simplicity, scalability, and good empirical success.
Several papers in this issue use variable ranking as a
baseline method (Caruana et al., 2003) (Weston et al.,
2003).
Let X a matrix containing m instances x
i
=
(x
i1
, . . . , x
id
) ∈ R
d
. we denote by y
i
= (y
1
, . . . , y
m
) the
vector of class labels for the m instances. A is the set
of features a
j
= (a
1
, . . . , a
d
).
Feature ranking makes use of a scoring function
H( j) computed from the values x
ij
and y
i
. By con-
vention, we assume that a high score is indicative of
a valuable variable and that we sort variables in de-
creasing order of H( j).
Feature ranking is a filter method thus it is inde-
pendent of the choice of the predictor. Even when
feature ranking is not optimal, it may be preferable
to other feature subset selection methods because of
its computational and statistical scalability: Compu-
tationally, it is efcient since it requires only the com-
putation of d scores and sorting the scores; Statisti-
cally, it is robust against overtting because it intro-
duces bias but it may have considerably less variance
(Hastie et al., 2001).
When applying scoring functions based on differ-
ent scoring criteria we have often different feature
ranking lists so finding a way to aggregate them is
required. The rank aggregation problem is to com-
bine many different rank orderings on the same set of
candidates, or alternatives, in order to obtain a bet-
ter ordering. This is a classical problem from social
choice and voting theory, in which each voter gives
a preference on a set of alternatives, and the system
outputs a single preference order on the set of alterna-
tives based on the voters’ preferences (Borda, 1781)
(Young, 1990).It is the key problem in many applica-
tions. For example, in sports and competition to rank
or to compare players from different eras. In machine
learning to do collaborative ltering and meta-search;
in database middleware to combine results from mul-
tiple databases. In recent years, rank aggregation
methods have emerged as an important tool for com-
bining information from different Internet search en-
gines or from different omics-scale biological studies
(Dwork et al., 2001). Ordered lists are routinely pro-
duced by today’s high-throughput techniques which
naturally lend themselves to a meta-analysis through
rank aggregation. (DeConde et al., 2006) proposed to
use rank aggregation methods to integrate the results
of several ordered lists of genes.
When aggregating feature rankings, there are two
issues to consider. The first one is which base fea-
ture rankings to aggregate. There are different ways
to generate the base feature rankings: the first uses
the same dataset, but by different ranking method, the
second one uses different subsamples of the dataset
but the same ranking method. In our experiments we
use the first ranking generation technique. The sec-
ond issue concerns the type of aggregation function
to use. There are mainly two kinds of rank aggre-
gation, score-based rank aggregation and order-based
rank aggregation. In the former, objects in the input
rankings are associated with scores, while in the latter,
only the order information of these objects is avail-
able. The Borda count (Borda, 1781) and median
rank aggregation are the most famous such method
where elements in the overall list are ordered accord-
ing to the average rank computed from the ranks in
all individual lists. Another category is based on the
majoritarian principles and attempts to accommodate
the ”majority” of individual preferences putting less
or no weight on the relatively infrequent ones. The
final aggregate ranking is usually based on the num-
ber of pairwise wins between items within individ-
ual lists. Any method that satisfies this condition,
known as the Condorcet criterion, is called the Con-
dorcet method (Young, 1990) (Young and Levenglick,
1978). In the recent literature, probabilistic models
on permutations, such as the Mallows model and the
Luce model, have been introduced to solve the prob-
lem of rank aggregation. In this work, we take advan-
tage from the two kinds, order-basedrank aggregation
but also score-based rank aggregation in order to not
lose the additional score information.
3 PROPOSED APPROACH
3.1 Optimization Problem
Rank aggregation provides a mean of combining in-
formation from different ordered lists and at the same
FeatureSelectionbyRankAggregationandGeneticAlgorithms
75
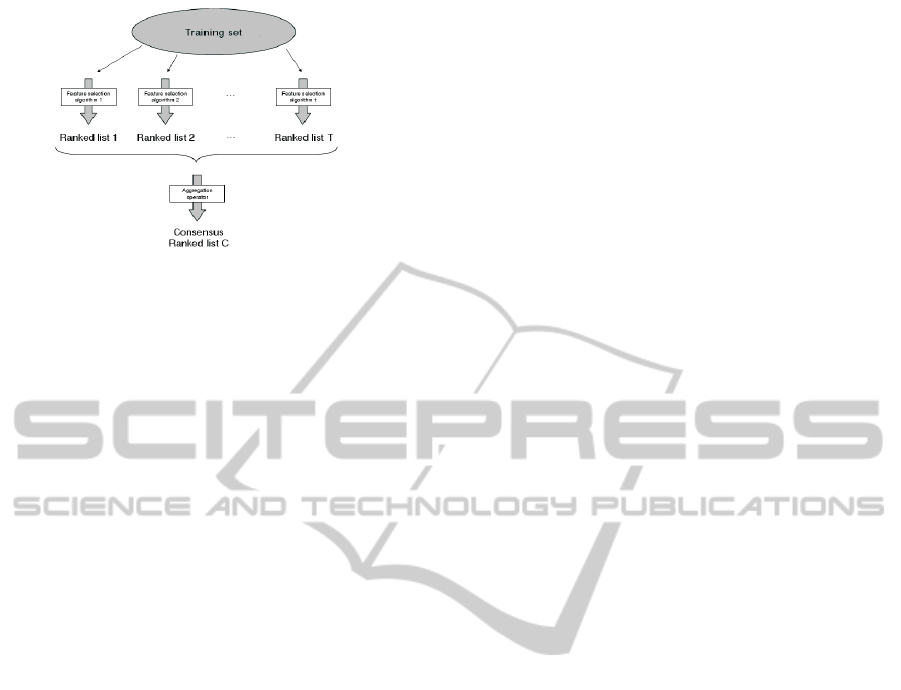
Figure 1: Rank Aggregation.
time, to set their weak points. The aim of rank aggre-
gation when dealing with feature selection is to find
the best list, which would be the closest as possible to
all individual ordered lists all together.
This can be seen as an optimization problem,
when we look at argmin(D, σ), where argmin gives
a list σ at which the distance D with a randomly se-
lected ordered list is minimized. In this optimization
framework the objective function is given by :
F(σ) =
m
∑
i=1
w
i
× D(σ, L
i
), (1)
where w
i
represent the weights associated with the
lists L
i
, D is a distance function measuring the dis-
tance between a pair of ordered lists ( for more details
see Section 3.2) and L
i
is the i
th
ordered list of cardi-
nality k. The best solution is then to look for σ
∗
which
would minimize the total distance between σ
∗
and L
i
given by
σ
∗
= argmin
m
∑
i=1
w
i
× D(σ, L
i
). (2)
3.2 Distance Measures
Measuring the distance between two ranking lists is
classical and several well-studied metrics are known
(Carterette, 2009; Kumar and Vassilvitskii, 2010), in-
cluding the Kendall’s tau distance and the Spearman
footrule distance. Before defining this two distance
measures, let us introduce some necessary notation.
Let S
i
(1), . . . , S
i
(k) be the scores coupled with the ele-
ments of the ordered list L
i
, where S
i
(1) is associated
with the feature on top of L
i
that is most important,
and S
i
(k) is associated with the feature which is at the
bottom that is least important with regard to the target
concept. All the other scores correspond to the fea-
tures that would be in-between, ordered by decreasing
importance.
For each item j ∈ L
i
, r( j) shows the ranking of this
item. Note that the optimal ranking of any item is 1,
rankings are always positive, and higher rank shows
lower preference in the list.
3.2.1 Spearman Footrule Distance
Spearman footrule distance between two given rank-
ings lists L and σ, is defined as the sum, overall the
absolute differences between the ranks of all unique
elements from both ordered lists combined. Formally,
the Spearman footrule distance between L and σ, is
given by
Spearman(L, σ) =
∑
f∈(L∪σ)
|r
L
( f) − r
σ
( f)| (3)
Spearman footrule distance is a very simple way for
comparing two ordered lists. The smaller the value of
this distance, the more similar the lists. When the two
lists to be compared, have no elements in common,
the metric is k(k + 1).
3.2.2 Kendall’s Tau Distance
The Kendall’s tau distance between two ordered rank
list L and σ, is given by the number of pairwise ad-
jacent transpositions needed to transform one list into
another (Dinu and Manea, 2006). This distance can
be seen as the number of pairwise disagreements be-
tween the two rankings. Hence, the formal definition
for the Kendall’s tau distance is:
Kendall(L,σ) =
∑
i, j∈(L∪σ)
K, (4)
where
K =
0 if r
L
(i) < r
L
( j), r
σ
(i) < r
σ
( j)
or r
L
(i) > r
L
( j), r
σ
(i) > r
σ
( j)
1 if r
L
(i) > r
L
( j), r
σ
(i) < r
σ
( j)
or r
L
(i) < r
L
( j), r
σ
(i) > r
σ
( j)
p if r
L
(i) = r
L
( j) = k + 1,
r
σ
(i) = r
σ
( j) = k + 1
(5)
That is, if we have no knowledge of the relative
position of i and j in one of the lists, we have sev-
eral choices in the matter. We can either impose
no penalty (0), full penalty (1), or a partial penalty
(0 < p < 1).
3.2.3 Weighted Distance
In case, the only information available about the in-
dividual list is the rank order, the Spearman footrule
distance and the Kendall’s tau distance are adequate
measures. However the presence of any additional
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
76

information about the individual list may improve
the final aggregation. Typically with filter methods,
weights are assigned to each feature independently
and then the features are ranked based on their rel-
evance to the target variable. It would be beneficial to
integrate these weights into our aggregation scheme.
Hence, the weight associated to each feature consists
of taking the average score across all of the ranked
feature lists. We find the average for each feature by
adding all the normalized scores associated to each
lists and dividing the sum by the number of lists.Thus,
the weighted Spearman’s footrule distance between
two list L and σ is given by
∑
f∈(L∪σ)
|W(r
L
( f)) × r
L
( f) −W(r
σ
( f)) × r
σ
( f)|.
=
∑
f∈(L∪σ)
|W(r
L
( f))−W(r
σ
( f))|×|r
L
( f)−r
σ
( f)|.
(6)
Analogously to the weighted Spearman’s footrule
distance, the weighted Kendall’s tau distance is given
by:
WK(L, σ) = |W(r
L
(i)) −W(r
L
( j))|K. (7)
3.3 Solution to Optimization Problem
Using Genetic Algorithm
The introduced optimization problem in Section 3.1
is a typical integer programming problem. As far as
we know, there is no efficient solution to such kind of
problem. One possible approach would be to perform
complete search. However, it is too time demanding
to make it applicable in real applications. We need to
look for more practical solutions.
The presented method uses a genetic algorithm for
rank aggregation. Genetic algorithms (GAs) were in-
troduced by (Holland, 1992) to imitate the mechanism
of genetic models of natural evolution and selection.
GAs are powerful tools for solving complex for solv-
ing combinatorial problems, where a combinatorial
problem involves choosing the best subset of com-
ponents from a pool of possible components in or-
der that the mixture has some desired quality (Clegg
et al., 2009). GAs are computational models of evo-
lution. They work on the basis of a set of candidate
solutions. Each candidate solution is called a ”chro-
mosome”, and the whole set of solutions is called a
”population”. The algorithm allows movement from
one population of chromosomes to a new population
in an iterative fashion. Each iteration is called a ”gen-
eration”. GAs in our case proceeds in the following
manner :
3.3.1 Initialization
Once a set of aggregation rank lists are generated by
several filtering techniques, it is necessary to create
an initial population of features to be used as starting
point for the genetic algorithm, where each feature
in the population represents a possible solution. This
starting population is then obtained by randomly se-
lecting a set of ordered rank lists.
Despite the success of genetic algorithm on a wide
collection of problems, the choice of the population
size stills an issue. (Gotshall and Rylander, ) proved
that the larger the population size, the better chance of
it containing the optimal solution. However, increas-
ing population size also causes the number of genera-
tions to convergeto increase. In order to havegreat re-
sults, the population size should depend on the length
of the ordered lists and on the number of unique el-
ements in these lists. From empirical studies, over a
wide range of problems, a population size of between
30 and 100 is usually recommended.
3.3.2 Selection
Once the initial population is fixed, we need to select
new members for the next generation. In fact, each el-
ement in the current population is evaluated on the ba-
sis of its overall fitness (the objective function score).
Depending on which distance is used, new members
(rank lists) are produced by selecting high performing
elements (Vafaie and Imam, 1994).
3.3.3 Cross-over
The selected members are then crossed-over with the
cross-over probability CP. Crossover randomly select
a point in two selected lists and exchange the re-
maining segments of these lists to create a new ones.
Therefore, crossover combines the features of two
lists to create two similar ranked lists
3.3.4 Mutation
In case only the crossover operator is used to produce
the new generation, one possible problem that may
arise is that if all the ranked lists in the initial popula-
tion have the same value at a particular rank, then all
future lists will have this same value at this particular
rank. To come over this unwanted situation a muta-
tion operator is used. Mutation operates by randomly
changing one or more elements of any list. It acts as a
population perturbation operator. Typically mutation
does not occur frequently so mutation is of the order
FeatureSelectionbyRankAggregationandGeneticAlgorithms
77

4 EXPERIMENTAL SETUP
4.1 Datasets
As discussed before using too much data, in terms of
the number of input variables, is not always effective.
This is especially true when the problem involves un-
supervised learning or supervised learning with un-
balanced data (many negative observations but mini-
mal positive observations). This paper addresses two
issues involving high dimensional data: The first is-
sue explores the behavior of ensemble method fea-
ture aggregation when analyzing data with hundreds
or thousands of dimensions in small sample size situ-
ations. The second issue deals with huge data set with
a massive number of instances and where feature se-
lection is used to extract meaningful rules from the
available data.
The experiments for the first case were conducted
on Central Nervous System (CNS), a large data set
concerned with the prediction of central nervous sys-
tem embryonal tumor outcome based on gene expres-
sion. This data set includes 60 samples containing 39
medulloblastoma survivors and 21 treatment failures.
These samples are described by 7129 genes(Pomeroy
et al., 2002). We consider also the Leukemia mi-
croarry gene expression dataset that consists of 72
samples which are all acute leukemia patients, ei-
ther acute lymphoblastic leukemia (47 ALL) or acute
myelogenous leukemia (25 AML). The total number
of genes to be tested is 7129. (Golub et al., 1999)
For the second case two credit datasets are used,
the German and the Tunisian credit dataset. The Ger-
man credit dataset covers a sample of 1000 credit con-
sumers where 700 instances are creditworthy and 300
are not. For each applicant, 21 numeric input vari-
ables are available .i.e. 7 numerical, 13 categorical
and a target attribute. The Tunisian dataset covers a
sample of 2970 instances of credit consumers where
2523 instances are creditworthy and 446 are not. Each
credit applicant is described by a binary target vari-
able and a set of 22 input variables were 11 features
are numerical and 11 are categorical. Table 2 displays
the characteristics of the datasets that have been used
for evaluation.
Table 1: Datasets summary.
Names German Tunisian CNS Leukemia
Instances 1000 2970 60 72
Features 20 22 7129 7129
Classes 2 2 2 2
Miss-values No Yes No No
4.2 Feature Rankers
We investigated three different filter selection algo-
rithms from the category of rankers, Relief algorithm
(Kira and Rendell, 1992), Correlation-based feature
selection (CFS) (Hall, 2000) and Information gain
(IG) (Quinlan, 1993). These algorithms are available
in Weka 3.7.0 machine learning package (Bouckaert
et al., 2009).
Relief algorithm evaluates each feature by its abil-
ity to distinguish the neighboring instances. It ran-
domly samples the instances and checks the instances
of the same and different classes that are near to each
other.
Correlation-based Feature Selection (CFS) looks
for feature subsets based on the degree of redundancy
among the features. The objective is to find the fea-
ture subsets that are individually highly correlated
with the class but have low inter-correlation. The sub-
set evaluators use a numeric measure, such as con-
ditional entropy, to guide the search iteratively and
add features that have the highest correlation with the
class.
Information gain (IG) measures the number of bits
of information obtained for class prediction by know-
ing the presence or absence of a feature.
4.3 Classification Algorithms and
Performance Metrics
We trained our approach using three well-known data
mining algorithms, namely Decision trees, Support
vector machines and The K-nearest-neighbor. These
algorithms are available in Weka 3.7.0 machine learn-
ing package (Bouckaert et al., 2009).
To evaluate the classification performance of each
setting and perform comparisons, we used several
characteristics of classification performance all de-
rived from the confusion matrix (Okun, 2011). We
define briefly these evaluation metrics.
The precision is the percentage of positive predic-
tions that are correct. The Recall (or sensitivity) is the
percentage of positive labeled instances that were pre-
dicted as positive. The F-measure can be interpreted
as a weighted average of the precision and recall. It
reaches its best value at 1 and worst score at 0.
The cited performance measures are obtained
when the cut-off is 0.5. However, changing this
threshold might modify previous results. In this pa-
per we use the AUC (the area under the ROC curve)
as graphical tool to evaluate the effect of selected fea-
tures on classification models (Ferri et al., 2009).
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
78
5 RESULTS
Tables 3-5 report on the performances achieved by
DT, SVM and KNN algorithms using the individ-
ual filters, mean aggregation and genetic algorithm.
In general the discussed feature selection approaches
perform well for selecting relevant features either for
small or high data dimensionality. There is obvi-
ously a strong similarity in the feature sets selected
by different approaches. A more detailed picture of
the achieved results shows that the precision of ag-
gregation approaches is better than most of the stud-
ied filters. Consistent with the theoretical analysis
for feature selection, the fusion approach usually out-
performs single filters. However, we should consider
individual results to confirm the superiority of our ap-
proach.
For information filtering as feature selection prob-
lems, one cares the most about both the precision and
the recall of the classification using individual and en-
semble feature selection methods. F-measure is the
metric of choice for considering both together.
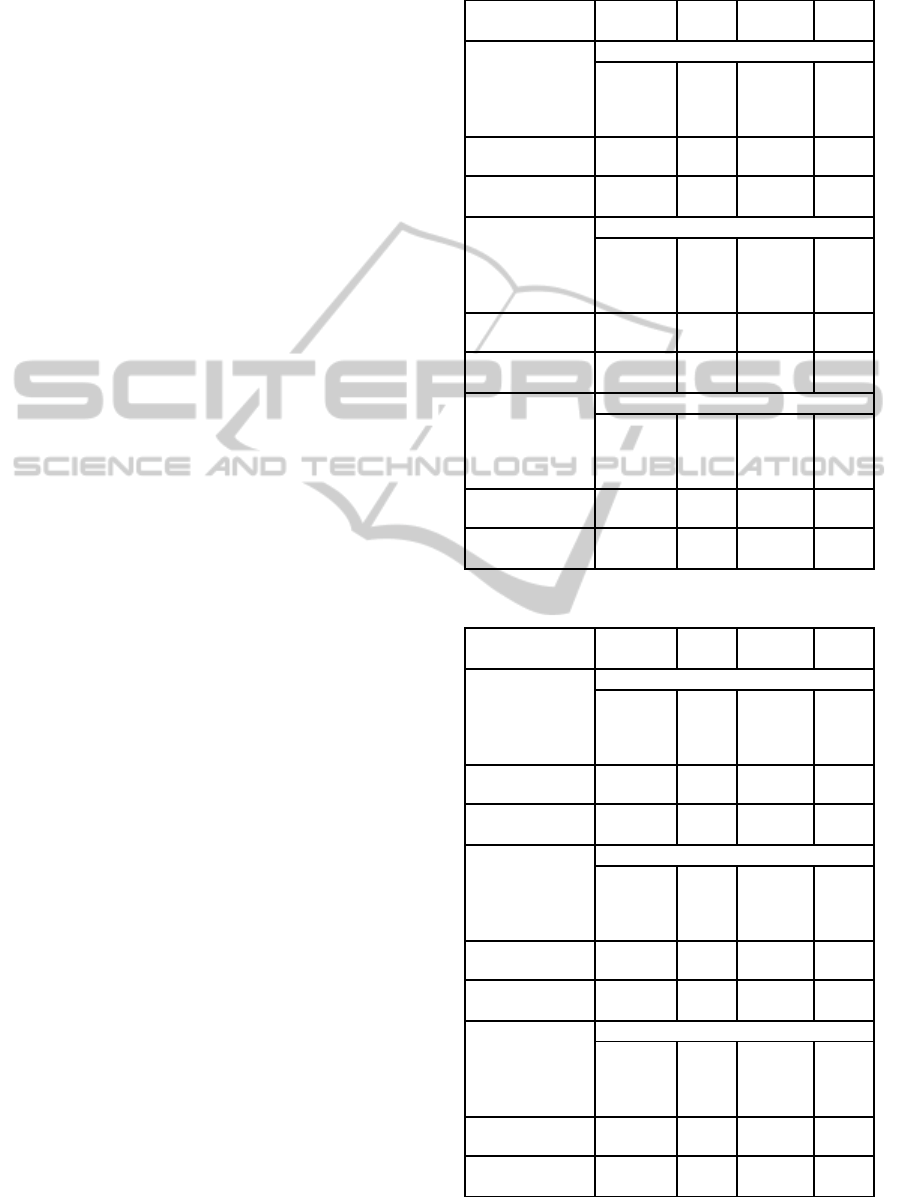
For the German dataset, in all cases the filters
aggregation gives better performance than individual
feature selection methods. For DT, the genetic al-
gorithm (GA) both with Kendall and Spearman dis-
tances gives better results than the mean aggregation
technique but it is not always true when taking ROC
area as a performance metric.
The outperformance of GA is also noticeable for
KNN classifier when comparing recall and F-measure
performance metrics. However for SVM classifier,
mean aggregation gives better results than GA. We
note also that for this dataset that GA either with
Kendall or Spearman distance functions gives the
same results.
Results of the Tunisian dataset given in Table 3
show that for DT classifier, the mean aggregation
technique outperforms GA aggregation. However,
when using SVM classifier, GA is better with com-
petitive results between the two distance functions.
GA with Spearman distance function is the best ag-
gregation technique with KNN classifier and this for
all performance criteria. Kendall and mean aggrega-
tion have similar results with a slight superiority of
precision and ROC area for mean aggregation tech-
nique.
For Leukemia dataset, GA with Kendall distance
gives the best results when a DT algorithm is trained.
It is followed by Spearman GA. Best results are also
given by Kendall for SVM classifier. Mean aggrega-
tion and Kendall give the same and best results with
KNN classifier.
Table 5 shows results of the CNS dataset. For this
Table 2: German dataset.
Precision Recall F-
Measure
ROC
Area
DT
Relief
0.709 0.73 0.707 0.706
Inf Gain 0.705 0.727 0.703 0.717
Correlation 0.695 0.717 0.697 0.715
Mean Aggreg
0.768 0.861 0.812 0.696
GA(Kendall) 0.773 0.883 0.824 0.709
GA(Spearman) 0.773 0.883 0.824 0.709
SVM
Relief
0.747 0.757 0.749 0.685
Inf Gain 0.746 0.756 0.748 0.684
Correlation 0.673 0.709 0.659 0.564
Mean Aggreg
0.782 0.884 0.83 0.654
GA(Kendall) 0.771 0.883 0.823 0.635
GA(Spearman) 0.771 0.883 0.823 0.635
KNN
Relief
0.698 0.707 0.701 0.691
Inf Gain 0.693 0.694 0.694 0.678
Correlation 0.701 0.712 0.705 0.714
Mean Aggreg
0.775 0.777 0.776 0.687
GA(Kendall) 0.771 0.797 0.784 0.683
GA(Spearman) 0.771 0.797 0.784 0.683
Table 3: Tunisian dataset.
Precision Recall F-
Measure
ROC
Area
DT
Relief
0.722 0.85 0.781 0.497
Inf Gain 0.814 0.852 0.809 0.547
Correlation 0.827 0.857 0.816 0.646
Mean Aggreg
0.865 0.981 0.919 0.56
GA(Kendall) 0.85 1 0.919 0.497
GA(Spearman) 0.863 0.983 0.919 0.558
SVM
Relief
0.769 0.847 0.784 0.5
Inf Gain 0.868 0.907 0.887 0.563
Correlation 0.769 0.847 0.784 0.505
Mean Aggreg
0.769 0.847 0.785 0.50
GA(Kendall) 0.85 1 0.919 0.5
GA(Spearman) 0.851 0.994 0.917 0.505
KNN
Relief
0.862 0.932 0.895 0.602
Inf Gain 0.86 0.94 0.898 0.607
Correlation 0.864 0.959 0.909 0.675
Mean Aggreg
0.864 0.938 0.899 0.644
GA(Kendall) 0.863 0.938 0.899 0.63
GA(Spearman) 0.866 0.941 0.902 0.645
FeatureSelectionbyRankAggregationandGeneticAlgorithms
79
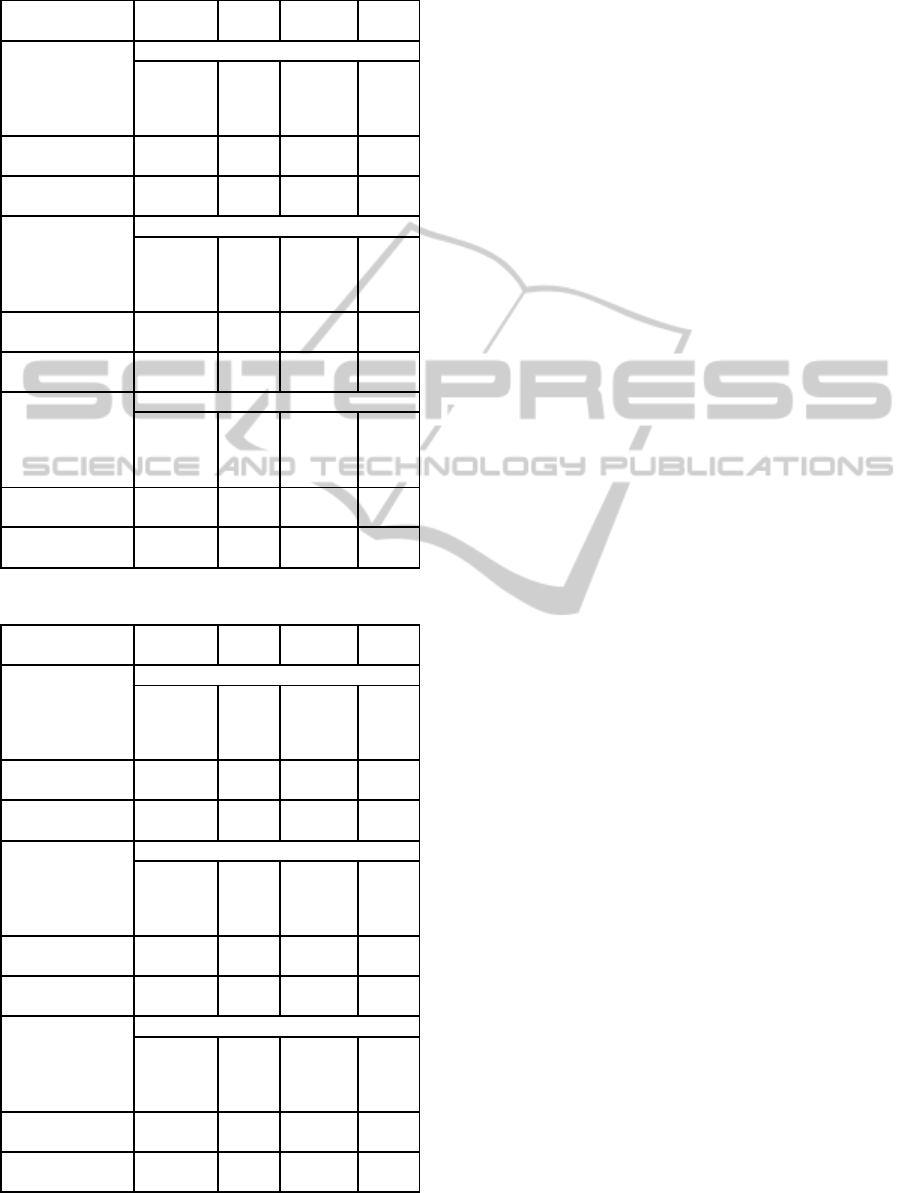
Table 4: Leukemia dataset.
Precision Recall F-
Measure
ROC
Area
DT
Relief
0.933 0.894 0.913 0.865
Inf Gain 0.913 0.894 0.903 0.871
Correlation 0.933 0.894 0.913 0.865
Mean Aggreg
0.951 0.83 0.886 0.866
GA(Kendall) 0.955 0.894 0.923 0.899
GA(Spearman) 0.952 0.851 0.899 0.878
SVM
Relief
0.972 0.972 0.972 0.969
Inf Gain 0.93 0.931 0.93 0.919
Correlation 0.958 0.958 0.958 0.949
Mean Aggreg
0.972 0.972 0.972 0.969
GA(Kendall) 0.986 0.986 0.986 0.98
GA(Spearman) 0.972 0.972 0.972 0.969
KNN
Relief
0.944 0.944 0.944 0.936
Inf Gain 0.92 0.917 0.917 0.92
Correlation 0.93 0.931 0.93 0.911
Mean Aggreg
0.973 0.972 0.972 0.951
GA(Kendall) 0.973 0.972 0.972 0.951
GA(Spearman) 0.958 0.958 0.958 0.938
Table 5: Central Nervous System dataset.
Precision Recall F-
Measure
ROC
Area
DT
Relief
0.600 0.538 0.568 0.399
Inf Gain 0.674 0.744 0.707 0.535
Correlation 0.676 0.641 0.658 0.512
Mean Aggreg
0.73 0.692 0.711 0.589
GA(Kendall) 0.795 0.795 0.795 0.74
GA(Spearman) 0.821 0.821 0.821 0.749
SVM
Relief
0.632 0.615 0.623 0.474
Inf Gain 0.737 0.718 0.727 0.621
Correlation 0.700 0.718 0.709 0.573
Mean Aggreg
0.825 0.846 0.835 0.756
GA(Kendall) 0.805 0.846 0.825 0.733
GA(Spearman) 0.875 0.897 0.886 0.83
KNN
Relief
0.659 0.692 0.675 0.513
Inf Gain 0.727 0.615 0.667 0.593
Correlation 0.677 0.538 0.600 0.531
Mean Aggreg
0.837 0.923 0.878 0.795
GA(Kendall) 0.787 0.949 0.86 0.736
GA(Spearman) 0.841 0.949 0.892 0.808
dataset, GA with Spearman distance function gives
very good results and outperforms GA with Kendall
distance and mean aggregation for the three classifi-
cation algorithms. Performance results of this algo-
rithm are especially excellent when using KNN clas-
sifier. GA with Kendall distance outperforms mean
aggregation for DT, but they have competitive results
for SVM and KNN classifiers. The performance im-
provement with all aggregation techniques comparing
to individual feature selection methods is very notice-
able for this dataset.
To summarize, we can notice that the achieved re-
sults show that the fusion performance is either supe-
rior to or at least as good as individual filter methods.
This confirms the theoretical assumption that rank ag-
gregation provides the means of combining informa-
tion from individual filtering methods and at the same
time overcome their weakness.
6 CONCLUSIONS
In this paper we present an overview of some of the
available ranking feature selection approaches. To get
the best results of all available alternatives, we com-
bine their results using rank aggregation. We demon-
strate that our rank aggregation algorithm can be used
to efficiently select important features based on differ-
ent filtering criteria. We effectively combine the ranks
of a set of filter methods via a weighted aggregation
that optimizes a distance criterion using genetic al-
gorithm. We illustrate our procedure using four real
datasets from biological and financial fields.
REFERENCES
Borda, J. C. D. (1781). Memoire sur les elections au scrutin.
Bouckaert, R. R., Frank, E., Hall, M., Kirkby, R., Reute-
mann, P., Seewald, A., and Scuse, D. (2009). Weka
manual (3.7.1).
Carterette, B. (2009). On rank correlation and the distance
between rankings. In Proceedings of the 32nd inter-
national ACM SIGIR conference on Research and de-
velopment in information retrieval, SIGIR ’09, pages
436–443, New York, NY, USA. ACM.
Caruana, R., Sa, V. R. D., Guyon, I., and Elisseeff, A.
(2003). Benefitting from the variables that variable se-
lection discards. jmlr, 3: 12451264 (this issue. pages
200–3.
Clegg, J., Dawson, J. F., Porter, S. J., and Barley, M. H.
(2009). A Genetic Algorithm for Solving Combina-
torial Problems and the Effects of Experimental Error
- Applied to Optimizing Catalytic Materials. QSAR
& Combinatorial Science, 28(9):1010–1020.
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
80

DeConde, R., Hawley, S., Falcon, S., Clegg, N., Knudsen,
B., and Etzioni, R. (2006). Combining results of mi-
croarray experiments: A rank aggregation approach.
Statistical Applications in Genetics Molecular Biol-
ogy, 5(1):1–17.
Dinu, L. P. and Manea, F. (2006). An efficient approach for
the rank aggregation problem. Theor. Comput. Sci.,
359(1):455–461.
Dwork, C., Kumar, R., Naor, M., and Sivakumar, D. (2001).
Rank aggregation methods for the web. pages 613–
622.
Ferri, C., Hern´andez-Orallo, J., and Modroiu, R. (2009).
An experimental comparison of performance mea-
sures for classification. Pattern Recognition Letters,
30(1):27–38.
Golub, T. R., Slonim, D.K., Tamayo, P., Huard, C., Gaasen-
beek, M., Mesirov, J. P., Coller, H., Loh, M. L., Down-
ing, J. R., Caligiuri, M. A., and Bloomfield, C. D.
(1999). Molecular classification of cancer: class dis-
covery and class prediction by gene expression moni-
toring. Science, 286:531–537.
Gotshall, S. and Rylander, B. Optimal population size and
the genetic algorithm.
Guyon, I. and Elisseff, A. (2003). An introduction to vari-
able and feature selection. Journal of Machine Learn-
ing Research, 3:1157–1182.
Hall, M. A. (2000). Correlation-based feature selection for
discrete and numeric class machine learning. In Pro-
ceedings of the Seventeenth International Conference
on Machine Learning, pages 359–366. Morgan Kauf-
mann.
Hastie, T., Tibshirani, R., and Friedman, J. (2001). The
Elements of Statistical Learning. Springer series in
statistics. Springer New York Inc.
Holland, J. H. (1992). Adaptation in natural and artificial
systems. MIT Press, Cambridge, MA, USA.
Kira, K. and Rendell, L. (1992). A practical approach to
feature selection. In Sleeman, D. and Edwards, P., ed-
itors, International Conference on Machine Learning,
pages 368–377.
Kohavi, R. and John, G. H. (1997). Wrappers for feature
subset selection. Artificial Intelligence, 97:273–324.
Kumar, R. and Vassilvitskii, S. (2010). Generalized dis-
tances between rankings. In Proceedings of the 19th
international conference on World wide web, WWW
’10, pages 571–580, New York, NY, USA. ACM.
Okun, O. (2011). Feature Selection and Ensemble Methods
for Bioinformatics: Algorithmic Classification and
Implementations.
Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M.,
Angelo, M., McLaughlin, M. E., Kim, J. Y. H., Goum-
nerova, L. C., Black, P. M., Lau, C., Allen, J. C.,
Zagzag, D., Olson, J. M., Curran, T., Wetmore, C.,
Biegel, J. A., Poggio, T., Mukherjee, S., Rifkin, R.,
Califano, A., Stolovitzky, G., Louis, D. N., Mesirov,
J. P., Lander, E. S., and Golub, T. R. (2002). Prediction
of central nervous system embryonal tumour outcome
based on gene expression. Nature, 415(6870):436–
442.
Quinlan, J. R. (1993). C4.5: programs for machine learn-
ing. Morgan Kaufmann Publishers Inc.
Vafaie, H. and Imam, I. (1994). Feature Selection Meth-
ods: Genetic Algorithms vs. Greedy-like Search.
Manuscript.
Weston, J., Elisseeff, A., Schlkopf, B., and Kaelbling, P.
(2003). Use of the zero-norm with linear models and
kernel methods. Journal of Machine Learning Re-
search, 3:1439–1461.
Young, H. P. (1990). Condorcet’s theory of voting. Math-
matiques et Sciences Humaines, 111:45–59.
Young, H. P. and Levenglick, A. (1978”,). A consistent ex-
tension of Condorcet’s election principle. SIAM Jour-
nal on Applied Mathematics, 35(2):285–300.
FeatureSelectionbyRankAggregationandGeneticAlgorithms
81
