
Involving End-users in Domain-Specific Languages Development
Experiences from a Bioinformatics SME
∗
Maria Jose Villanueva, Francisco Valverde and Oscar Pastor
PROS Research Center, Universitat Politècnica de València, 46022 Valencia, Spain
Keywords:
Software Engineering, Domain-Specific Languages, Agile Development.
Abstract:
Involving end-users in software development is a goal envisioned by the Software Engineering community.
As they have the domain knowledge, it is feasible to develop software applications that really fulfil their
requirements. Domain-specific languages (DSL) are widely applied to accomplish this vision. However,
end-users collaboration in DSL development is also important to ensure that their needs are well understood
and represented. This research work proposes a DSL development process that combines methodological
guidelines for DSL development with good practices from agile methods to encourage end-user involvement.
In this paper, we overview the complete process and we focus on the two first stages: Decision and Analysis.
In order to illustrate the proposal, it is applied in the development of a DSL for a bioinformatics SME that
works on genetic disease diagnosis.
1 INTRODUCTION
The importance of the end-user involvement in soft-
ware development has been a topic widely discussed
in the academia (Ko et al., 2011; Fischer et al., 2004).
The role of end-user experts in any development pro-
cess is essential, especially in complex domains as
bioinformatics, where most of the domain knowledge
is difficult to be translated by developers to a suitable
implementation.
Domain-specific languages (Fowler, 2010) have
been proposed as a solution to reduce the knowledge
gap between end-users (or domain experts) and devel-
opers. According to Spinellis (Spinellis, 2001), some
of their benefits are: 1) the concrete expression of do-
main knowledge; 2) the direct involvement of the do-
main expert in the software life cycle; and 3) the better
expressiveness to describe domain implementations.
According to Van Deursen et al. (Van Deursen et al.,
2000), DSLs can abstract the underlying software im-
plementation as a set of domain concepts that end-
users can easily understand. If an executable DSL is
provided, end-users can use it to write their own pro-
grams.
Current DSL development methodologies (Strem-
beck and Zdun, 2009; Ceh et al., 2011) take into ac-
count end-users in requirements gathering and in de-
ployment. Usually, developers create DSLs from a re-
∗
Small and Medium Enterprise
quirements specification without additional end-users
participation. As a consequence, the discovery of any
domain misunderstanding is delayed until a first ver-
sion is delivered. This fact substantially increases the
development time of the DSL if, for instance, the lan-
guage editor or the execution environment must be
reimplemented accordingly.
Agile Software Development (Highsmith and
Cockburn, 2001) and the Agile Manifesto (Beck et al.,
2001) have praised for a set of good practices to im-
prove software development. Among these practices,
they have encouraged the involvement of end-users
(or stakeholders) throughout the project, and the short
delivery of prototypes for early error detection. We
believe that some of these practices can be profitable
in the context of DSL development.
The contributions of this research work are: 1) the
identification of good practices from agile methods
that apply in the DSL development context; and 2) the
inclusion of these practices inside a DSL development
process to improve end-user involvement.
Our DSL development process is based on the
methodological guidelines proposed by Mernik et al.
(Mernik et al., 2005)—to define the different pro-
cess stages and activities— and the agile methods
XP (Beck and Andres, 2004), Scrum (Schwaber and
Beedle, 2002) and Agile Modeling (Ambler, 2002)—
to analyse different agile practices. Briefly, the pro-
cess is configured to create the DSL incrementally by
97
Villanueva M., Valverde F. and Pastor O..
Involving End-users in Domain-Specific Languages Development - Experiences from a Bioinformatics SME.
DOI: 10.5220/0004450000970108
In Proceedings of the 8th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE-2013), pages 97-108
ISBN: 978-989-8565-62-4
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

means of small releases that satisfy a set of require-
ments. End-users participate in some activities during
the development of each release—so they can provide
constant feedback— and also evaluating the resulting
DSL release implementation—so any detected error
can be fixed in the next iteration.
In this paper we provide an overview of the pro-
cess and we focus on the two first stages: Decision
and Analysis. To illustrate both stages, we describe
the development of a DSL for genetic disease diagno-
sis, which has been elaborated in close collaboration
with experts from a bioinformatics SME. Due to the
complexity of this evolving domain, this real scenario
highlights the need of sound methodological practices
to support a good communication with domain ex-
perts.
The rest of the paper is structured as follows. Sec-
tion 2 reviews other approaches that research how to
involve end-users or how to use agile practices in DSL
development. Section 3 overviews the proposal: first,
the methodological base for DSL development (pro-
posed by (Mernik et al., 2005)); second, the agile
practices that apply in DSL context; and third, the ag-
ile DSL development process proposed. Section 4 and
5 describe in detail the Decision and Analysis stages,
respectively, and several examples from a DSL for ge-
netic disease diagnosis illustrate each stage. Section
6 discusses the main findings while applying the pro-
posal. Finally, section 7 explains the conclusions and
the future work.
2 RELATED WORK
Agility and end-user involvement in DSLs develop-
ment have been previously addressed by the Software
Engineering community.
On the one hand, several authors have addressed
end-users involvement within software development
processes.
The authors of (Pérez et al., 2011) propose a new
method that encourages the analysis of end-users role
in the development process. This approach takes
into account good practices of end-user development,
analyses user-requirements and develops the DSL ac-
cordingly. As a result, their method generates a visual
domain-specific language by means of a tool. This ap-
proach adopts end-user development good practices
to improve DSL development, which better satisfy
user needs, with the aim to involve them afterwards
in the future Model-Driven development process. On
the contrary, our approach adopts good practices from
agile methods to promote end-user involvement dur-
ing the DSL development and to ensure their prefer-
ences are fulfilled within the DSL.
Also, the authors of (Izquierdo and Cabot, 2012)
provide a collaborative infrastructure for their in-
volvement in every stage of DSL development. Using
their proposal end-users and developers interact un-
til artefacts of each stage satisfy end-users’ require-
ments. This approach supports an active involvement
of end-users in all the stages of the development pro-
cess. On the contrary, our approach promotes the use
of agile practices as a way to make end-users involve-
ment easier, but avoids their participation in the def-
inition of artefacts outside their domain knowledge,
such as conceptual models, which sometimes may be
useless, if not detrimental.
On the other hand, several authors have addressed
the application of agile principles on software devel-
opment processes.
The authors of (Grigera et al., 2012) propose to
bridge agile practices with model-driven development
of Web applications by using Test-Driven Develop-
ment. Starting with a set of user stories, authors
propose the use of graphical user interface mock-
ups for improving stakeholders involvement and re-
quirements gathering. Authors also introduce a DSL,
WebSpec, for specifying the interaction and naviga-
tion requirements. Following an agile iterative cycle
and model-driven principles, a WebML model is de-
rived that represents a Web application. Our approach
shares some agile practices from this proposal, but
it is mainly focused on DSL development instead of
software generation from requirements.
Also, the work of (Visser, 2008) proposes the use
of good practices from agile methods in DSLs devel-
opment, such as carrying out the DSL development
incrementally, addressing one concern at a time. This
approach addresses the design and implementation of
the DSL from abstractions obtained after analysing
domain technologies, instead from a previous domain
analysis. Likewise, our approach applies agile prac-
tices for DSL development and adopts an iterative cy-
cle. However, our approach follows a deductive pro-
cess (where a domain analysis is used in the design
and this design is also used in the implementation)
and promotes end-users involvement to improve their
satisfaction about requirements coverage.
3 OVERVIEW OF AN AGILE DSL
DEVELOPMENT PROCESS
Mernik et. al. discusses (Mernik et al., 2005) when
and how to develop a DSL and provides a set of guide-
lines for future developers. The authors describe a
development process made up of five stages: Deci-
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
98
sion, Analysis, Design, Implementation and Deploy-
ment; and several patterns that aid developers in each
of them. In a recent work (Ceh et al., 2011), they have
extended the process to seven stages adding Testing
and Maintenance. This approach has been widely ap-
plied in different DSL development projects, for ex-
ample (Jr. et al., 2011; Arora et al., 2009; Visser,
2008). Because of these reasons, we have selected
this work as a base of our proposal.
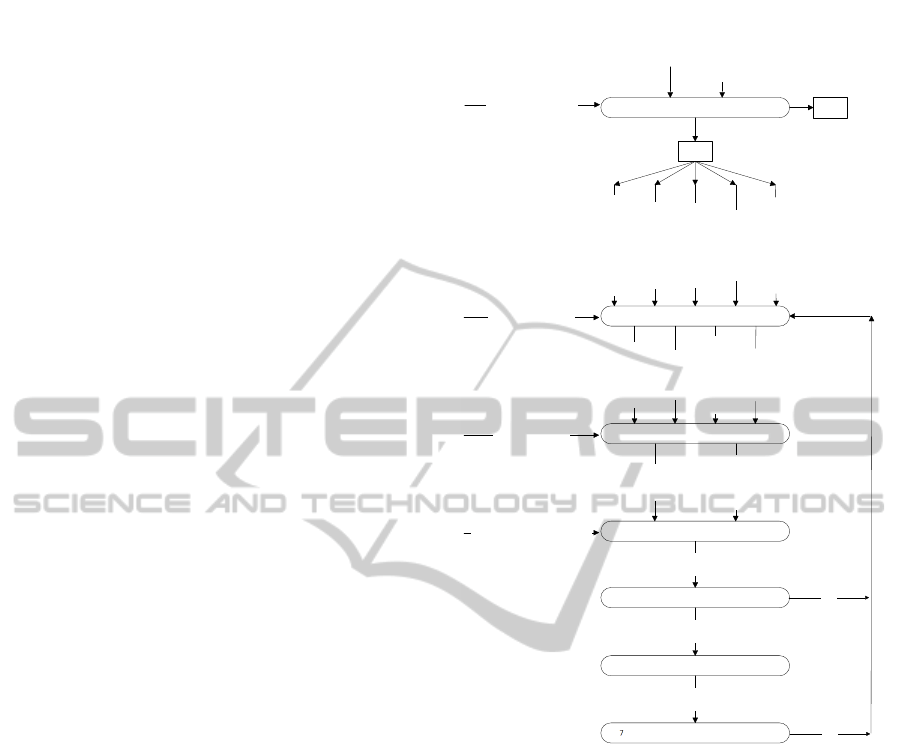
Figure 1 (taken and adapted from (Ceh et al.,
2011)) depicts the stages—with the corresponding in-
puts and outputs— and the proposed patterns for the
the first four stages: First, a Decision (1) is made
about developing or not the DSL taking into account
domain experts demands and the current state of the
domain; If so, an Analysis (2) is performed,using dif-
ferent assets that represent the domain, to obtain a set
of formal artefacts that represents the domain. Once
the domain is clearly represented it is time to Design
(3) the target DSL constructs; those designs are used
to create an executable Implementation (4); which, in
order to identify its correctness, is the subject of a
Testing (5); if everything is correct, a Deployement (6)
is carried out to be used by end-users and, optionally,
a Maintenance (7) will perform any required change.
Agile methods have become widely popular for
achieving end-user involvement and improving the
time-to-market of software products (Beck et al.,
2001). Next, we explain the practices selected from
XP, Scrum and Agile Modeling suitable for DSL de-
velopment:
• Architectural Envisioning. This practice, pro-
posed by the Agile Modeling approach, encour-
ages the early identification of a viable technical
strategy. Many times DSL development is guided
by domain concepts, and how the DSL is going
to be executable is decided in late stages. Be-
cause of that, sometimes is difficult to translate
concepts to a working implementation, as some
authors have stated (Visser, 2008). Applying this
practice, developers can select and adapt domain
concepts according to the DSL execution environ-
ment expected.
• User Stories. In order to manage requirements
in agile methods, they are divided in user stories,
which are brief descriptions related by end-users
about a demand that contributes to add value. The
set of user stories provides a simplified view of
the functional features to be developed. In the
context of DSL development, user stories are an
effective instrument to discover the language con-
structs and concepts to be introduced in the DSL.
Hence, user stories guide the development and
provide traceability between initial requirements
Ontology Driven Development of Domain-Specific Languages
ComSIS Vol. 8, No. 2, Special Issue, May 2011
321
specification, attribute grammars, denotational semantics, or algebraic
specifications for semantic specification).
Decision
Analysis
Design
Implementation
Deployment
Maintenance
Technical literature
Existing
implementations
Custom surveys
Expert advice
Current and future
requirements
Terminology
Concepts
Commonalities
Variations
Syntax
Semantics
DSLDSL
Possible
existing
implementations
Requirements
Yes
No
Testing
DSL
DSL
Fig. 1. DSL development phases
Implementation. Different approaches for DSL development can be used,
such as: interpreter, compiler/application generator, embedding,
(2)
(3)
(4)
(5)
(6)
( )
(1)
DSL
Domain analysis
method
Implementation pattern
i.e. Interpreters,
Compilers/application
generators, Emdedding, ...
Informal, Formal (DSSA,
FODA, ODM), Extract from
code
Design pattern
Language exploitation,
Language invention
Decission pattern
i.e. Notation, AVOPT,
Task automation, Product
line, ...
Figure 1: Original DSL development process(Ceh et al.,
2011).
and the final DSL constructs. Also, user stories
are useful to aid end-users to establish their prior-
ities about the requirements to be introduced into
the DSL.
• Acceptance Tests or Usage Scenarios. In order
to check the fulfilment of requirements in agile
methods, end-users briefly describe scenarios that
must be accomplished by the software to be devel-
oped. In DSL development such scenarios are in-
teresting not only for testing purposes but also for
improving the understanding about the domain.
In our approach, acceptance tests are the primary
validation source to ensure that the DSL satisfies
end-users’ expectations.
• Iteration Planning or Sprint. The majority of ag-
ile methods manage the development in a set of
iterations in which only a small set of features is
implemented. This practice encourages faster re-
InvolvingEnd-usersinDomain-SpecificLanguagesDevelopment-ExperiencesfromaBioinformaticsSME
99

leases that can be evaluated by stakeholders, or
end-users in our context. DSL development can
benefit from this practice by checking if the con-
cepts required by end-users are included in incre-
mental subsets of the DSL. Also, errors can be
detected without developing the complete DSL in-
frastructure (editor, execution environment, etc.).
• Incremental Design. Together with a development
based on iterations, agile practices promote the
incremental design of the software (models, UI,
components, etc.). Many DSL development ap-
proaches design the whole abstract language from
a previous domain analysis. The design of a full
DSL is a time-consuming task and it is not fin-
ished until end-users evaluate the result. We be-
lieve that if DSL developers and end-users are fo-
cused on an small set of constructs or user stories,
DSL can be built more easily and it is more flexi-
ble to changes.
Our work proposes an agile DSL development
process that combines the aforementioned method-
ological guidelines with the chosen agile practices,
to provide faster small releases of the DSL and to
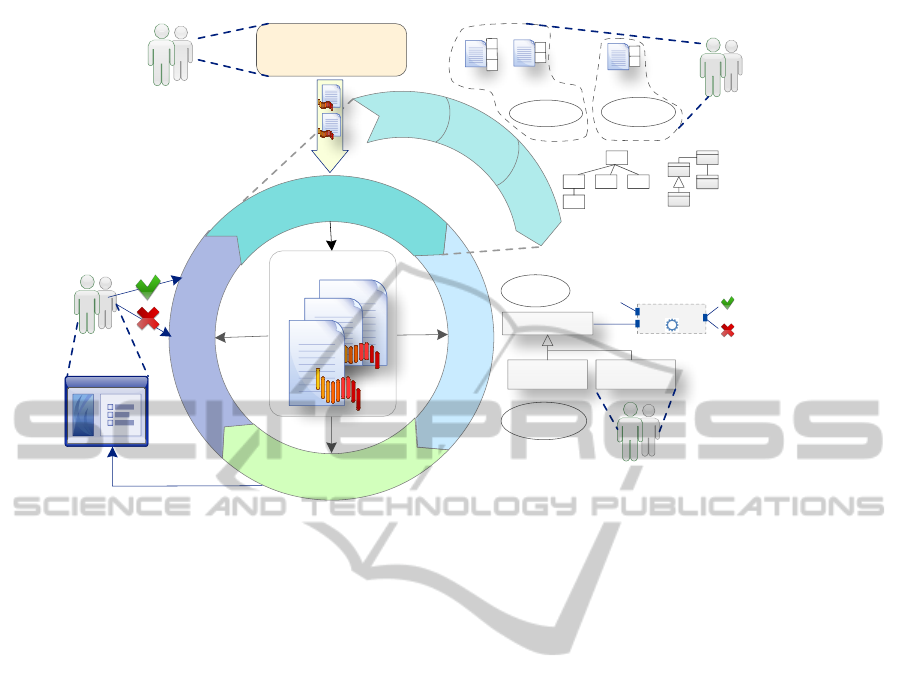
involve end-users. Figure 2 overviews the proposal.
The most relevant feature of the process is the in-
clusion of an iterative cycle for the stages Analy-
sis, Design, Implementation and Testing. The stages
Deployment and Maintenance have been avoided for
simplification purposes.
In the Decision stage all decision patterns—
Notation, AVOPT, Task automation, Product line,
Data structure representation, Data structure traver-
sal, System front-end, Interaction and GUI construc-
tion from Mernik et. al guidelines—are adopted to
decide if developing the DSL is worth. End-users pro-
vide information about existing tools and about their
requirements. Following the agile practice architec-
tural envisioning, we propose to make a deeper use
of this information to elaborate a review (1) where
available technology such as DSLs, software tools,
frameworks, etc., are assessed regarding end-users’
requirements. This review is useful to better justify
the decision to develop the DSL and, eventually, to
make early decisions about the architecture and the
implementation platform.
After this stage, following the agile practices it-
eration planing and incremental design, the original
sequential process, which includes Analysis, Design,
Implementation and Testing, is transformed into an
iterative process. Decision Stage remains outside be-
cause the decision to design the DSL is only made
once.
The proposed iterative cycle gathers:
• Analysis Stage. Following the agile practices user
stories, acceptance tests and iteration planning,
this stage is extended with two steps:
– Requirements Specification (2). This step re-
fines the preliminary requirements specification
detailing all requirements expected for a com-
plete DSL. To comply with agile practices, end-
user’s requirements are divided in a set of user
stories (2a) and acceptance tests (2b), in a sim-
ilar way than XP and Scrum.
– Iteration Planning (3). This step completes the
specified user stories with their priorities and
settles a list (or sprint backlog using the Scrum
terminology) that defines their implementation
order according to end-users’ preferences. In
order to optimize the iterative cycle, in each it-
eration a set of user stories are chosen to be im-
plemented, instead of addressing one user story
per iteration. An agreed release deadline is
scheduled and an acceptance test (3a) that cov-
ers all these user stories is defined.
– Domain Analysis (4). This step follows the in-
formal pattern from Mernik et al. guidelines to
create a set of artefacts that precisely describe
the DSL domain. We choose this pattern as
a way to avoid the overloading that carrying
out a complete formal method may introduce.
We adopt the definition of a feature model (4a)
(firstly introduced by (Kang et al., 1990)) to ex-
press the variabilities and commonalities of the
DSL, and the definition of a conceptual model
(4b) (a UML Class Diagram) to precisely de-
scribe the terminology, the concepts involved
in the DSL and the relations among them. To
comply with the agile practice incremental de-
sign, these artefacts are created incrementally,
so they represent only the sub-domain related
with the selected user stories of the current iter-
ation (and the previous ones).
Due to end-users are more interested in talking
about their domain problems than working in soft-
ware development issues that address them (Fis-
cher et al., 2009), we have promoted their partic-
ipation in steps directly related with such prob-
lems: In the Requirements Specification step they
explain their requirements and tests to be satisfied,
and in the Iteration Planning step they point out
their priorities and specify a scenario that gath-
ers several user stories. On the contrary, their in-
volvement in the development of feature and class
models is avoided. In our experience, end-users
are not comfortable with this kind of models, and
the lack of understanding could create mistakes
(Costabile et al., 2008).
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
100
Im p lem entation
T
e
s
t
i
n
g
D
e
s
i
g
n
A n alysis
en d-users
D ecisio n
Re q uirem en ts
Techn o lo g ical
re v ie w
en d-use rs
S e m a n tic s
S y n ta x
A b stra ct S y n ta x
C o re Syn ta x
(te xtual)
E n d -U ser
S y ntax (vis u a l)
Requirem en ts
Spec ificatio n
I
t
e
r
a
t
i
o
n
P
l
a
n
n
i
n
g
D
o
m
a
i
n
A
n
a
l
y
s
i
s
en d-use rs
U se r
Story 3
t1
t2
U se r
Story 2
t1
t2
U se r
Story 1
t1
t2
t3
A cce p ta n ce
te st
A cce p ta n ce
te st'
Ite ra tio n x Ite ra tio n x '
Variab ilitie s/
Com m on alitie s
D om ain
Conce p ts
Sce na rio
i
Ite ra tio n i
Te sts
i
Sto rie s
i
en d-use rs
Valid a tio n
t
i
Pro to ty pe
2
2a
2b
3
4
3a
4a 4b
5
5a
5b
1
1a
6
6a
7
7a
Figure 2: Overview of the agile DSL development process proposed.
• Design Stage (5). Following the agile practice in-
cremental design, the syntax and semantics de-
signs represent only the user stories of the cur-
rent iteration (and the previous ones). As an op-
tional step, end-users can express their prefer-
ences about the end-user oriented (concrete) syn-
tax (5a), which normally is a visual projection of
the core (concrete) textual syntax. Additionally,
developers validate that the Abstract syntax can be
instantiated to support all acceptance tests speci-
fied in the Analysis stage (5b).
• Implementation Stage (6). Following the agile
practices incremental design and architectural en-
visioning, the DSL is implemented taking into ac-
count the partial syntax and semantics from the
Design stage and the technological review per-
formed in the Decision stage. As a result, a pro-
totype (6a) DSL that complies with the set of user
stories (from this iteration and the previous ones)
is created (or evolved from previous iterations).
End-users are not involved in this stage.
• Testing Stage (7): In this stage the prototypical
implementation is tested against the requirements
from the current iteration. First, the specified ac-
ceptance tests are checked. And then, the pro-
totype is shown to end-users so they check the
acceptance tests and provide their opinion (7a):
agreeing with the functionality they expected and
rejecting the functionality they don’t like. Addi-
tionally, end-users can provide insights about new
requirements they are interested in.
After the last stage, if there are some requirements
left, a new iteration is started. Because the Testing
stage captures feedback from end-users, requirements
may have been updated, removed, added or changed
their priority, so next iteration may need to perform
some adjustments. Concretely, all these changes are
addressed in the Requirements Specification and Iter-
ation Planning steps from the Analysis stage. Since
requirements and priorities are revisited each itera-
tion according to end-users feedback, the develop-
ment planning is constantly adapted to satisfy their
ongoing necessities.
4 DECISION STAGE
In this section, we apply the Decision stage of our
proposal to develop a DSL for customizing genetic
disease diagnosis software. First, we explain a real
scenario in the genetics domain where geneticists face
a set of problems related with end-user software de-
velopment. As a solution, we propose the develop-
ment of a DSL and we explain how it fits to solve
their problems. Second, we provide a preliminary de-
scription of geneticists’ requirements, a review about
technologies for the development of genetic disease
diagnosis software and the suitable decision patterns
that apply within the context of the DSL.
InvolvingEnd-usersinDomain-SpecificLanguagesDevelopment-ExperiencesfromaBioinformaticsSME
101

4.1 Genetic Disease Diagnoses
In the genetics domain, DNA structures and their
relationships with human traits are too difficult and
heterogeneous to be understood by people outside
the domain. For this reason, geneticists with some
programming knowledge have been traditionally de-
veloping their own databases, analysis algorithms,
scripts, frameworks and development tool-kits over
the years.
Even so, as not every geneticist is experienced
in software development, they create scientific work-
flows to reuse available software applications. Unfor-
tunately, these software applications are often created
to fulfil specific requirements rather than being de-
signed for reuse. Customization and data exchange, if
possible, often requires advanced programming skills
that geneticists don’t usually have.
A common scenario to face this problem is that
geneticists create spread sheets to perform data oper-
ations and copy results into text files themselves to
exchange data among different applications. This sit-
uation highly impacts on their productivity, and it has
become even more critical since the recent improve-
ments in sequencing technologies, which now pro-
duce more genetic data than experts are able to anal-
yse (Rusk, 2009).
An example of this situation is in IMEGEN
(IMEGEN, ), a bioinformatics SME working on the
diagnosis of genetic diseases. In order to understand
its context, Figure 3 overviews its process for genetic
disease diagnosis:
1. Patient’s Variation Gathering Phase: A patient’s
DNA sample is inspected with a software tool
(chosen according to the diagnosis technique per-
formed) to obtain a report of genetic variations.
The term variation is used to express how the con-
tent of the patient’s DNA in a specific position
is different from the content of a “disease-free”
DNA reference sequence in the same position.
2. Disease Knowledge Phase: Each variation de-
tected in the previous phase is searched in differ-
ent databases and websites until success. If a vari-
ation has been previously described, additional in-
formation related to a genetic disease is retrieved.
If not, it is characterized as an unknown variation.
3. Report Generation Phase: All genetic variations
and their associated information are gathered in a
report according to geneticists’ preferences. To do
this, they use a spread sheet or a word-document
template to represent all the information.
If the analysis outcome is only a single gene, ge-
neticists from IMEGEN are able to perform this pro-
R e p o rt G e n era tio n
V a ria tion G a the rin g D ise a se K n o w led g e
D
n
a
S
a
m
p
l
e
A lig n m en t
T o o l
S a m p le
A n aly sis
G D B
V a ria tions
K
n
o
w
l
e
d
g
e
D ete cte d
V ariatio n s
R ep o r t
U n k n o w n
V aria tio ns
C
l
a
s
s
i
f
i
c
a
t
i
o
n
B iblio grap y
& D ise a se
1
2
3
C G H A rray
T o o l
R e sults
K n o wn
V aria tio ns
D ise as e
V aria tio ns
U n h arm fu l
V aria tio ns
...
Figure 3: IMEGEN’s Genetic Disease Diagnosis Process.
cess manually: interacting with different tools and
searching information through different databases.
However, when they must manage bigger amounts of
genetic data and take into account a wider array of
knowledge resources (websites, databases, scientific
papers, etc.), their current procedure is no longer fea-
sible.
Geneticists from IMEGEN need to create their
own software applications that support different ge-
netic disease diagnoses, but the software mechanisms
to do it must avoid programming issues and hide tech-
nological details. In this context, a DSL is a good
choice to provide such mechanisms.
Working with IMEGEN, we have realized that the
genetic disease diagnoses observed share the same
process, but also, each disease diagnosis introduces
slight variations. For example, both Medullary Thy-
roid Cancer and Achondroplasia diagnoses require
identifying genetic variations that geneticists asso-
ciate with the target disease. However, the Cancer
is analysed with a technology that provides the pa-
tient’s sequence in SEQ format, while the technology
for the Achondroplasia provides the patient’s varia-
tions in VCF format. Once obtained the variations, for
the cancer, only the exons (regions) 10 and 11 from
the gene RET are validated with different databases
while for the Achondroplasia, locating the variations
“p.Gly380Arg” or “p.Gly375Cys” in the gene FGFR3
is enough to confirm the disease. All these common-
alities and variabilities that are directly related with
the genetics domain are well understood by geneti-
cists so they are perfect candidates to become con-
structs of the DSL.
In summary, geneticists can be provided with a
“programming language” for specifying genetic dis-
ease diagnoses. This new language will provide ab-
stractions at the genetic domain knowledge level, that
is, related to genetic concerns that geneticists under-
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
102

stand, in order to manage the underlying technologi-
cal implementation.
4.2 A DSL for Customizing Genetic
Disease Diagnosis Software
With this goal in mind, we address the definition of
a DSL to compose and customize genetic disease
diagnosis software. The DSL will support mono-
genic and multigenic disease diagnoses. At the mo-
ment, among all kinds of diseases, the DSL ad-
dresses the ones where a cause-effect relation can
be established between a genetic variation and a
disease (an example list can be downloaded from
http://www.imegen.es/cms_genetic_diagnosis.php).
A brief summary of the requirements they expect
to create a customized tool are:
• R1. Retrieve all variations from a patient once
its sample has been analysed chemically: because
there are different techniques and technologies
that use different outputs, this requirement is de-
composed in three:
– R1.1 Get variations from a file that contains the
patient’s sequence data.
– R1.2 Get variations from a file that contains the
alignment between the patient’s sequence and a
reference sequence.
– R1.3 Get variations from a file that contains the
patient’s variations.
• R2. Search the patient’s variations in differ-
ent genetic databases: each analysis will require
a different set of genetic databases, sometimes
databases that gather knowledge from different
diseases and others disease-specific databases.
• R3. Filter the list of patient’s variations for a spe-
cific region or look for a set of interesting varia-
tions: when a disease is being diagnosed not every
variation is always relevant.
• R4. Gather in a report only the essential informa-
tion for the target disease: using their expertise,
geneticists choose the meaningful variations and
the proper knowledge related with them.
Now, end users enumerate the tools they know in their
domain to “create” themselves their own software ap-
plications. Following our approach, we review these
tools to evaluate the support of the named require-
ments. Our goals are to assess if the current state of
the domain technology satisfies geneticists’ expecta-
tions, to detect if a DSL is useful in the domain, and to
identify what could be reused for its implementation
environment.
Table 1: Technological Support of Geneticists Require-
ments.
Biojava Taverna Galaxy Alamut
R1.1 Partially No No No
R1.2 Partially No Yes No
R1.3 No No Yes Yes
R2 No Partially Partially Yes
R3 No Partially Partially No
R4 No Yes No Partially
Some examples of these tools are: Biojava (ver-
sion 3.0.5) (Holland et al., 2008), Taverna (version
2.4) (Hull et al., 2006), Galaxy (online version)
(Goecks et al., 2010) and Alamut (version 2.2) (Ala-
mut, ). Table 1 summarizes the coverage of the afore-
mentioned requirements by the suggested tools.
Biojava is a framework, based on the Java lan-
guage, composed by a set of programming libraries
to create Java programs for analysing genetic data.
Biojava supports the reading of several file formats
and the execution of analysis algorithms; however,
in order to combined them to identify the patient’s
variations, geneticists should have Java programming
skills. The rest of requirements are not supported.
Taverna is a desktop tool for the design, edition
and execution of workflows based on the integra-
tion of web services specially focused on the biolog-
ical domain. It allows retrieving data from databases
that comply with Biomart technology (Haider et al.,
2009), and saving results in XML or XLS format.
However, some programming knowledge is required
to search variations inside the data obtained by re-
trieval services and to filter them. It does not support
to identify variations from patient’s data.
Galaxy is a web environment that integrates a
set of genetic services and allows running those ser-
vices independently or creating workflows combining
them. It integrates services that combined, eventually,
are able to identify variations from alignments or im-
port a list of variations, retrieve data from a set of ge-
netic databases and filter the obtained data sets. How-
ever, it requires a deeper knowledge of Galaxy struc-
ture and programming knowledge to identify varia-
tions from patient’s sequence data and search patient’s
variations in genetic databases. It does not support to
create customized reports.
Alamut is a desktop tool that provides a user-
friendly interface for the interpretation of genetic
variations. Variations can be retrieved by reading a
file that contains a list or adding them, one by one,
manually. Once in the system, variations can be
searched into a set of predefined genetic databases.
However, in order to create customized reports users
must program templates and perform post processing
InvolvingEnd-usersinDomain-SpecificLanguagesDevelopment-ExperiencesfromaBioinformaticsSME
103

operations. It is not possible to read variations from
the patient’s sequence or alignments, neither to filter
variations.
This technological review envisions the need for
more streamlined and easier to use software cus-
tomization mechanisms, because although almost all
requirements can be satisfied by the analysed tools,
geneticists are forced to learn a programming lan-
guage and deal with technological details (XML con-
figuration files, workflow design, database connec-
tions, etc.) to fully accomplish their necessities.
Mostly, they are reluctant to learn a new technology
because they are not sure if the time invested is worth.
A DSL will hide these low-level details and
its technological implementation will integrate some
functionalities of these tools under higher level con-
structs related with diagnosis concerns. For exam-
ple, from the reviewed tools it can be reused: 1) file
readers from Biojava; 2) genetic data retrieval mecha-
nisms from Taverna, Galaxy or Alamut; 3) filters from
Galaxy; and 4) file writers from Taverna or Alamut.
After the technological review of bioinformatic
tools, we go on with the decision. Among all deci-
sion patterns adopted from the original methodology,
we consider that the following ones justify the deci-
sion to proceed with the DSL development:
1. Task Automation. The DSL can automate a set
of activities that geneticists are performing man-
ually. For example, nowadays geneticists are re-
sponsible for the interoperability among software
applications, a difficult task due to the wide array
of genetic data formats available. Thus, a set of
DSL constructs will manage the format interoper-
ability among different software artefacts.
2. Product Line. The DSL can be used to config-
ure highly similar genetic disease diagnoses that
conform to the general process. Geneticists will
express the specific properties depending of the
disease and the diagnosis approach.
3. Interaction. The DSL can hide technological de-
tails that are outside the scope of geneticists’
knowledge. For example, geneticists are respon-
sible to discern the most suitable DNA alignment
algorithm from a wide array of choices. This
should be decided using genetic and diagnosis
features instead of algorithm-related implementa-
tions. For example, geneticists should choose be-
tween “protein alignment” and “nucleotide align-
ment” instead of between “blastp” (blast algo-
rithm for proteins) and “blastn” (blast algorithm
for nucleotides) algorithms.
4. GUI Construction. The DSL can be used to cus-
tomize how geneticists interact with data. They
will be able to describe their own interfaces by
indicating the inputs they want to provide to the
diagnosis and the reports to be generated.
5 ANALYSIS STAGE
In this section, we apply the Analysis stage of our
proposal to develop a DSL for customizing genetic
disease diagnosis software. We explain the Require-
ments Specification, Iteration Planning and Domain
Analysis steps of the first iteration and provide a set
of examples.
5.1 Requirements Specification
In this step, the preliminary requirements specifica-
tion is extended with the elaboration of user stories
and acceptance tests.
As we have discussed in Section 3, user stories
and acceptance tests are the agile practices selected
for requirements specification. In order to describe
user stories, agile methods encourage the use of few
sentences written by end-users avoiding the use of dif-
ficult technical syntax. These sentences usually de-
scribe the users role, the action to be accomplished
and the goal they pursue. A set of examples from our
DSL for are:
1. User story 1 (associated with requirement R1.1):
“I want choose a file with SEQ format from the
computer, so that the patient’s sequence is read”.
2. User story 2 (associated with requirement R1.1):
“I want choose a reference sequence from the
computer, so that it can be used afterwards to per-
form comparisons”.
3. User story 3 (associated with requirement R1.1):
“I want to get a reference sequence from the NCBI
database by indicating its RefSeq identifier, so
that it can be used afterwards to perform compar-
isons”.
4. User story 4 (associated with requirement R1.1):
“I want to align a patient sequence against a refer-
ence sequence, so that I can see how they match”.
5. User story 5 (associated with requirement R1.3):
“I want to choose a file with VCF format from
the computer, so that the patient’s variations are
read”.
6. User story 6 (associated with requirement R2):”I
want to search the variations against the dbSNP
database, so that patient’s variations that are SNPs
can be identified”.
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
104

7. User story 7 (associated with requirement R3):
“I want to indicate a list of variations in
HGVS_Protein notation, so that only those vari-
ations are taken into account”.
For each user story, several acceptance tests are de-
fined. An acceptance test is a description of usage
(real inputs and expected outputs) of a user story that
is defined with the aim to check the correctness of
its implementation. When a user story is delivered, it
can only be considered complete after it has passed
all its acceptance tests. These tests are different from
unit tests because they check the functionality taking
into account end-user’s experience, nor the expected
behaviour of internal implementation details. In order
to describe them, it is encouraged the same guidelines
than user stories, to use of few sentences written by
end-users that describe the user role, the action they
will be performed within the system and the response
they expect. The acceptance tests related with the user
story 5 are:
1. Acceptance Test 5.1: “I will choose the file
3Variations.vcf to check that the three varia-
tions {g.1234A>C, g.4567C>G, g.8910insAA}
are read”.
2. Acceptance Test 5.2: “I will choose the file
Empty.vcf and I expect to get an error that says
that the file is empty”.
3. Acceptance Test 5.4 “I will choose the file NoRef-
erence.vcf and I expect to get an error that says
that the reference has not been indicated”.
4. Acceptance Test 5.3: “I will choose the file Vari-
ations.sam and I expect to get an error that says
that the file has the wrong format”.
The major effort invested by end-users in this step is
performed in the first iteration. In the following it-
erations, end-users will refine those user stories and
acceptance test that, for any reason, were incorrect or
unambiguous, remove the ones that are not required
anymore and include the new ones.
5.2 Iteration Planning
In this step, end-users prioritize user stories and
choose the ones that are implemented in the current
iteration. In each iteration, end-users define one or
more acceptance test that gather several user stories.
These acceptance tests, which we name scenarios, are
different from previous acceptance tests because they
are designed to check user stories correctness work-
ing as a unit. An scenario is a reflection for end-users
about what they will be able to accomplish with the
prototypical implementation at the end of the itera-
tion.
In our example, in the first iteration, end-users to-
gether with developers have established that the most
important user stories are the number 5 and 7 because:
1) they are the most easier and simpler to be trans-
lated into a DSL; and 2) they provide a remarkable
improvement over their current practices. An exam-
ple of scenario that gathers both user stories is:
Scenario 1: “To diagnose the Achrondoplasia Dis-
ease, I will chose the file “variations.vcf” (which
I obtained from my sequencing machine in the
VCF format) to see if the patient has the variations
p.Gly375Cys and p.Gly380Arg”
This acceptance test reflects what geneticists ex-
pect to accomplish at the end of the first iteration: 1)
To provide a VCF file that is saved in their computer
(user story 5); 2) to see the variations once the file
is selected (user story 5); 3) to provide a list of ex-
pressions in HGVS_Protein notation (user story 7);
and 4) to see only the variations that match in their
HGVS_Protein notation with any of the expressions
provided (user story 7).
5.3 Domain Analysis
The goal of this step is to represent unambiguously
the domain of the DSL. Our process adopts the in-
formal pattern proposed by the original methodology
and proposes the definition of a feature model and a
conceptual model (as explained in section 3).
According to the agile practice incremental de-
sign, the domain to be represented by both models
corresponds only to the set of user stories addressed
in the current iteration. Because both models are in-
crementally created in each iteration, this step adds
new elements, such as attributes or relationships, or
refines the existing ones.
Following with our example, the models that rep-
resent the user stories 5 and 7 are:
• The Feature Model (Figure 4) has as a root feature
Genetic Diagnosis, which represents the type of
applications that can be expressed using the DSL.
Taking into account the addressed user stories,
these applications gather two capabilities, which
are represented in the model as two branches:
– Patient Data (left branch): Represents the in-
formation of a patient analysed to diagnose a
genetic disease. In the current iteration, the
only kind of patient’s data is a set of Variations
expressed in the VCF format. This branch rep-
resents the capabilities related to the user story
5.
– Variation Analysis (right branch): Represents
the activities to be performed in order to iden-
tify any evidence of a disease in a patient. In
InvolvingEnd-usersinDomain-SpecificLanguagesDevelopment-ExperiencesfromaBioinformaticsSME
105

VCF
Genetic Diagnosis
Patient Data Variation Analysis
Variations
Filter
Optional
Mandatory
HGVS_Protein
Figure 4: Feature Model from Iteration One.
the current iteration, this analysis consists of
applying a Filter over the list of patient’s Vari-
ations. The filter criteria is expressed using
the HGVS_Protein notation. This branch rep-
resents the features related to the user story 7.
We must remark that all features explained in this
model are mandatory because in this first itera-
tion user stories do not describe any alternative
feature. It is expected that next iterations will pro-
vide new features that will reveal: new branches,
optional features, or new choices under a feature.
For example, there is a user story that requires fil-
tering the variations using the HGVS_Coding no-
tation. To represent this user story, a new child of
Filter will be defined and the mandatory property
of HGVS_Protein will be changed for a choice be-
tween the two notations.
• The Conceptual Model (Figure 5) gathers the
main concepts involved in genetic disease diag-
noses: Disease, Patient and Variation:
– The entity Patient represents a human being
that has taken a genetic test to find out if he
or she is genetically predisposed to get a cer-
tain disease. It has an attribute id that identifies
a patient from others. This entity represents a
concept of the user story 5.
– The entity Variation describes the content of the
patient’s DNA in a specific position or region,
but it is called variation because it is expressed
in regards of a reference DNA. To perform the
diagnosis, geneticists are interested in the Vari-
ations that a Patient has, represented by a com-
position association between both entities. The
attribute HGVS_ProteinNotation is a notation
created by the genetics community to express
unambiguously what is occurring in the patient
at the protein level (which protein should be
created and which one is being created by the
patient). The entity and the relationship repre-
-HGVS_ProteinNotation
Variation
-Name
Disease
1 *
1
*
predisposes
-id
Patient
has
Figure 5: Conceptual Model from Iteration One.
sent the concepts involved in the user story 5,
and the attribute HGVS_ProteinNotation rep-
resents a concept involved in the user story 7.
– The entity (Genetic) Disease is a condition re-
lated with the DNA chemicals that causes a
body malfunction. The attribute name identi-
fies the disease inside the medical community.
A diagnose is made when it is found a relation-
ship between a patient’s Variation and a Dis-
ease, which predisposes the patient to get the
disease. This relationship is made thanks to ge-
neticists knowledge about the disease. The en-
tity, attribute and association represent the con-
cepts involved in the user story 7.
6 DISCUSSION
After applying the two first stages described to de-
velop a DSL for a bioinformatics SME, we have
found that combining methodological guidelines for
DSL development with agile practices improves end-
user involvement in several ways. This feedback has
been gathered in the meetings with two geneticists,
three bioinformaticians and two developers—while
carrying out the Decision stage and the Analysis stage
of the first iteration of the process.
First, end-users are able to understand the artefacts
which they are required to interact and contribute. For
example, they use their own words to specify user sto-
ries and acceptance tests. This fact is important be-
cause a good communication between end-users and
DSL developers reduces misunderstanding about end-
users’ requirements and aids in the detection of errors.
Second, end-users are able to lead the process and
make decisions. The ongoing process can be adapted
to their immediate necessities because they are re-
sponsible to decide which user stories must be accom-
plished in each iteration. This situation encourages
end-users to actively participate because they feel rel-
evant for the DSL development.
Additionally, we have found that end-users ac-
quire a better knowledge of the current state of the
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
106

DSL development process. As they participate in
the creation of the iteration planning, they are aware
about what user stories have been already accom-
plished, what is being addressed and when user stories
are planned to be finished. This situation improves the
relationship between end-users and developers: end-
users know exactly what to expect from developers
and they do not create themselves false expectations
that lead to future disappointments.
We should remark that these facts are not thor-
oughly validated as we have detected them during the
first iteration planning. In future work, we plan to
carry out more iterations to support these findings and
also to asses the complete process. For example, ask-
ing end-users about their opinions regarding the diffi-
culties found, the time they invested and the support
to react against their changing needs.
In summary, the benefits observed up now show
promising results of the use of good practices from
agile methods in DSL development to involve end-
users.
7 CONCLUSIONS
This research work proposes an agile DSL develop-
ment process that adopts a set of practices from agile
methods, with the main goal of improving end-users
involvement in DSL development. The agile practices
included are: 1) an iterative cycle, which addresses
only a set of end-users’ requests each iteration; 2) a
requirements specification activity based on the spec-
ification of user stories and acceptance tests, which
ensures that end-users communicate their ongoing re-
quirements in each iteration; and 3) an iteration plan-
ning step based on priorities and scenarios, which en-
sures that end-users express their ongoing priorities
and expectations. Thanks to these agile practices, a
better involvement of end-users in DSLs development
is expected as the preliminary results show.
We must remark that end-users involvement sup-
poses an overloading for some of them: they do not
appreciate the direct benefits or they are only inter-
ested in the finished DSL.This fact highlights the ne-
cessity of an agile DSL development process, which
is our main goal.
As future work, we will detail the stages not in-
cluded in this paper: Decision, Implementation and
Testing. Our goal is to clearly define each stage, se-
lecting the agile practices that improve end-user in-
volvement. Simultaneously, we will continue with the
development of the DSL for genetic disease diagnosis
in order to observe the practical benefits. Addition-
ally, geneticists from the SME will test the final DSL
with different disease diagnosis in order to assess the
completeness of the DSL developed.
ACKNOWLEDGEMENTS
The authors would like to thank IMEGEN for all
these years of collaboration, providing both useful
genetic knowledge and a real environment for re-
search, and also GEMBiosoft, especially to Dr. Ana
M. Levin, for its support in the development of
this paper. This work has been developed with the
support of MICINN under the FPU grant AP2009-
1895, the project PROS-Req (TIN2010-19130-C02-
02), and co-financed with ERDF.
REFERENCES
Alamut. Interactive biosoftware.
Ambler, S. (2002). Agile modeling: Effective practices.
John Wiley and Sons.
Arora, R., Mernik, M., Bangalore, P., Roychoudhury, S.,
and Mukkai, S. (2009). A domain-specific language
for application-level checkpointing. In Proceedings of
the 5th International Conference on Distributed Com-
puting and Internet Technology, ICDCIT ’08, pages
26–38, Berlin, Heidelberg. Springer-Verlag.
Beck, K. and Andres, C. (2004). Extreme programming
explained: embrace change. Addison-Wesley Profes-
sional.
Beck, K., Beedle, M., van Bennekum, A., Cockburn, A.,
Cunningham, W., Fowler, M., Grenning, J., High-
smith, J., Hunt, A., Jeffries, R., et al. (2001). The agile
manifesto. http://agilemanifesto.org/principles.html
Accesed 2013, 7(08).
Ceh, I., Crepinšek, M., Kosar, T., and Mernik, M. (2011).
Ontology driven development of domain-specific lan-
guages. Computer Science and Information Systems,
8(2):317–342.
Costabile, M., Mussio, P., Parasiliti Provenza, L., and Pic-
cinno, A. (2008). End users as unwitting software
developers. In Proceedings of the 4th international
workshop on End-user software engineering, pages 6–
10. ACM.
Fischer, G., Giaccardi, E., Ye, Y., Sutcliffe, A., and
Mehandjiev, N. (2004). Meta-design: a manifesto for
end-user development. Communications of the ACM,
47(9):33–37.
Fischer, G., Nakakoji, K., and Ye, Y. (2009). Metadesign:
Guidelines for supporting domain experts in software
development. Software, IEEE, 26(5):37–44.
Fowler, M. (2010). Domain-specific languages. Addison-
Wesley Professional.
Goecks, J., Nekrutenko, A., Taylor, J., and Team, T. (2010).
Galaxy: a comprehensive approach for supporting ac-
cessible, reproducible, and transparent computational
research in the life sciences. Genome Biol, 11(8):R86.
InvolvingEnd-usersinDomain-SpecificLanguagesDevelopment-ExperiencesfromaBioinformaticsSME
107

Grigera, J., Rivero, J., Robles Luna, E., Giacosa, F., and
Rossi, G. (2012). From requirements to web applica-
tions in an agile model-driven approach. Web Engi-
neering, pages 200–214.
Haider, S., Ballester, B., Smedley, D., Zhang, J., Rice, P.,
and Kasprzyk, A. (2009). Biomart central portal: uni-
fied access to biological data. Nucleic acids research,
37(suppl 2):W23–W27.
Highsmith, J. and Cockburn, A. (2001). Agile software de-
velopment: The business of innovation. Computer,
34(9):120–127.
Holland, R., Down, T., Pocock, M., Prli
´
c, A., Huen, D.,
James, K., Foisy, S., Dräger, A., Yates, A., Heuer, M.,
et al. (2008). Biojava: an open-source framework for
bioinformatics. Bioinformatics, 24(18):2096–2097.
Hull, D., Wolstencroft, K., Stevens, R., Goble, C., Pocock,
M., Li, P., and Oinn, T. (2006). Taverna: a tool for
building and running workflows of services. Nucleic
acids research, 34(suppl 2):W729–W732.
IMEGEN. Instituto de medicina genomica.
Izquierdo, J. and Cabot, J. (2012). Community-driven lan-
guage development. MiSE’12.
Jr., I. F., Fister, I., Mernik, M., and Brest, J. (2011). De-
sign and implementation of domain-specific language
easytime. Computer Languages, Systems and Struc-
tures, 37(4):151–167.
Kang, K., Cohen, S., Hess, J., Novak, W., and Peter-
son, A. S. (1990). Feature-Oriented Domain Analysis
(FODA) Feasibility Study. Technical report.
Ko, A. J., Abraham, R., Beckwith, L., Blackwell, A., Bur-
nett, M., Erwig, M., Scaffidi, C., Lawrance, J., Lieber-
man, H., Myers, B., Rosson, M. B., Rothermel, G.,
Shaw, M., and Wiedenbeck, S. (2011). The state of
the art in end-user software engineering. ACM Com-
puting Surveys (CSUR), 43(3):21:1–21:44.
Mernik, M., Heering, J., and Sloane, A. M. (2005). When
and how to develop domain-specific languages. ACM
Computing Surveys (CSUR), 37(4):316–344.
Pérez, F., Valderas, P., and Fons, J. (2011). Towards the
involvement of end-users within model-driven devel-
opment. End-User Development, pages 258–263.
Rusk, N. (2009). Focus on next-generation sequencing data
analysis. Nature Methods, 6(11s):S1.
Schwaber, K. and Beedle, M. (2002). Agile software de-
velopment with Scrum, volume 18. Prentice Hall PTR
Upper Saddle Riverˆ eNJ NJ.
Spinellis, D. (2001). Notable design patterns for domain-
specific languages. Journal of Systems and Software,
56(1):91–99.
Strembeck, M. and Zdun, U. (2009). An approach
for the systematic development of domain-specific
languages. Software: Practice and Experience,
39(15):1253–1292.
Van Deursen, A., Klint, P., and Visser, J. (2000). Domain-
specific languages: An annotated bibliography. ACM
Sigplan Notices, 35(6):26–36.
Visser, E. (2008). Webdsl: A case study in domain-specific
language engineering. Generative and Transforma-
tional Techniques in Software Engineering II, pages
291–373.
ENASE2013-8thInternationalConferenceonEvaluationofNovelSoftwareApproachestoSoftwareEngineering
108
