
Computational Biology Modeling across Different Scales
Filippo Castiglione
1
and Francesco Pappalardo
2
1
Istituto per le Applicazioni del Calcolo, National Research Council of Italy, Viale Manzoni 30, 00185 Rome, Italy
2
Dipartimento di Scienze del Farmaco, University of Catania, Viale A. Doria 6, 95125 Catania, Italy
Keywords: Mathematical Biology, Computational Methods, Multi-Scale Models, Systems Biology.
Abstract: One of the most formidable challenges in modern biology is to get a unified view of the various
mechanisms governing the behavior and of the causal relationships among different parts of a living system.
It is coming clearer nowadays that to get such comprehensive picture computational models embracing
different observation levels in space and time have to be formulated to explain the enormous amount of data
deriving from -omic high throughput measurements methods. In this article we aim at giving a meaning to
the concept of multi-scale modeling in the framework of studies of biological systems with particular
interest in understanding human physiology in disease conditions.
1 INTRODUCTION
Mathematical models of natural phenomena intend
to describe reality. By means of the mathematical
formalism allowing logical reasoning over
designated variables we account for observations
made through experimentation. Defining the
variables of a mathematical model is a fundamental
step actually setting up the range of logical
deductions allowed by that model. For example, if
we use a variable to describe the changes of a
concentration of a protein in the blood we are
definitely overlooking the dynamics of the atoms
and the ions hence we cannot get any information
about the folding of the protein itself. The origin of
this oversight lays in a basic principle sometimes
referred to as the lex parsimoniae most commonly
known as the Ockam’s Razor. “Pluralitas non est
ponenda sine neccesitate” in very simple words
states that in the description of a phenomenon the
most useful model is the most parsimonious one in
terms of elements used. In this regard, following up
the example above, it makes little sense to describe
the laws governing the forces accounting for the
folding of the protein if we are interested in the half-
life of the protein and we can estimate its decay rate
by fitting a curve to a set of experimental data about
the concentration in the blood of that protein.
William of Ockham was a Franciscan monk and
logician who lived in the 14
th
century in a village of
the English county of Surrey. At that time the
principle of parsimony in describing and modeling a
natural phenomena was well reasoned. However
today, the situation is a “bit” different. The lex
parsimoniae is still valid and indeed very much used
when describing a phenomenon, but besides
classical mathematical models allowing for an exact
analytical approach, another modus operandi is now
commonly employed. This is what we can call the
synthetic approach consisting in constructing a
replica or toy of the studied system in terms of the
most important identified elements and the laws
governing the relationship among them. Actually
this approach is not new at all. The “engineer”
Leonardo used to construct toy models of flight
machines before attempting anything real-scale.
What is new today is that we can use digital
computers to construct toy models. We can instruct
extremely powerful CPUs to execute algorithms
representing entities and laws and we can then make
all kinds of conceptual experiments on those entities
and laws. This “digital synthetic” approach is
commonly referred to as simulation. Today, when
studying a certain natural phenomena, scientists first
identify elements and basic laws governing the
dynamics of the system, then they represent them as
data structures and algorithms and finally execute
the algorithms to observe how the system evolves.
The Ockam’s principle is still valid and used in the
first phase of this process but after that, thanks to the
fact that computers do the calculations, the
parsimony is forsaken, and the complexity of the
617
Castiglione F. and Pappalardo F..
Computational Biology Modeling across Different Scales.
DOI: 10.5220/0004405706170625
In Proceedings of the 3rd International Conference on Simulation and Modeling Methodologies, Technologies and Applications (BIOMED-2013), pages
617-625
ISBN: 978-989-8565-69-3
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
initial toy model is augmented by simply adding
new entities and laws. Indeed, with little difficulty
we can detail processes incorporating hypothetical
or experimentally derived knowledge. We can even
compose pre-constructed models of different parts of
the real system or arrange models describing reality
at different scales of observation. This holistic
approach is what in biology is called systems biology
(Kitano, 2002). The class of systemic models
therefore includes the one of multi-scale models.
Multi-scale modeling has been drawn a great
deal of attention in biological modeling and is
discussed in many recent articles and reviews (Qu et
al., 2011; Dada and Mendes, 2011; Southern et al.,
2008; Bassingthwaighte et al., 2005; Coveney and
Fowler, 2005; Engler et al., 2009; Grima, 2008). See
for example the interesting attempt to provide a
framework for multi-scale computational modeling
that is given in (Sloot and Hoekstra, 2010) together
with two examples showing how to bridge different
single-scale models.
The present article aims at giving a meaning to
the concept of multi-scale modeling in the
framework of studies of biological systems in
general with particular interest in understanding
human physiology in disease conditions. This article
provides a general introduction to the
methodological issues of multi-scale modeling. For a
more extensive reading including examples we
recommend the above-cited reviews and also
(Hunter and Nielsen, 2005; Meier-Schellersheim, et
al., 2009; Murtola et al., 2009; Schnell et al., 2007;
Bradley et al., 2011; Joshi et al., 2011).
2 LEVELS OF BIOLOGICAL
ORGANIZATION
Before we define what a multi-scale models is, it is
first necessary to make clear what it is meant to
formulate a model at a certain scale (Southern et al.,
2008). In the natural sciences, to make an
observation requires setting a temporal and a
dimensional scale. For example, freely draw from
disparate scientific fields, the phenomena of the
continental drift is better described over a time span
of million years, the evolution of a disease like
multiple sclerosis in years or decades, the immune
recognition of an infectious agent in days, the cell
cycle and circadian rhythm in twenty-four hours and
so on, to fast processes like the heart beat lasting
about a second or the fold of a protein that takes
place in microseconds and beyond. Likewise, certain
phenomena are better seen over a length or space
scale of light years, as for example the formation of
galaxies, or kilometers, like for the propagation of a
tsunami wave, or micrometers to describe the
duplication of a cell, and so on.
In general terms, while we can intuitively say if a
determined process involves cells, molecules, or
organs, it is not so simple to identify values for the
lengths at which we switch from one level to the
next (Southern et al., 2008). Major levels of
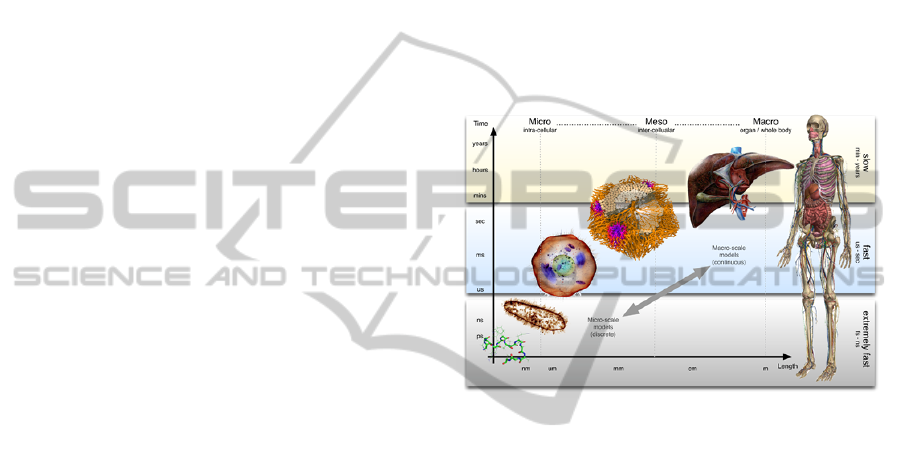
biological organization are regulated at scales of
many orders of magnitude in space and time
(Figure), with space spanning from the molecular
scale (10
-10
m) to the living organism scale (1 m), and
time from nanoseconds (10
-9
s) to years (10
8
s).
Figure 1: Multi-scale models of the human body targeting
complex processes span many time and length scales of
biological organization.
When combining models in a systemic way, the
challenge remains in the manner the components are
put together. Note that, in the study of complex
phenomena as for instance human pathologies, a
unified view is indeed necessary to reach a
comprehension of the various mechanisms in action
and of the causal relationships among different parts
of that complex system that is the human body (Di
Ventura et al., 2006). Complex diseases entail
phenomena ranging all scales, from observations at
the microscopic scales (from pico to micro meters)
to microscopic tissue damage and embracing
temporal events ranging from very fast processes
lasting in the order of femto seconds (for example
protein folding, protein docking, etc.) to slower
microscopic events like DNA transcription, cellular
mechanisms like meiosis or even lengthy ones like
the embryogenesis or the evolution of a disease like
diabetes or cancer (Hunter and Borg, 2003). In this
regard, there is another important aspect that should
not be left out from the whole picture. This is the
contemporary data explosion deriving from
genomic, transcriptomics, proteomics and
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
618
metabolomics studies consisting in high dimensional
datasets produced by latest high throughput
measurements methods (Deane et al., 2002). Also,
other types of data coming from modern microscopy
and biological imaging contribute to the detailed
description of the constitutive parts and basic
structures of living organisms (Southern et al.,
2008). On that account, the current challenge
expects to relate these datasets to higher level
phenotypic characteristics and computational multi-
scale modeling approaches are set to reveal
quantitative mechanistic relationships between these
various measurements (Di Ventura et al., 2006). For
example, high throughput gene expression data can
be used to infer knowledge of the intracellular
activities that can be later ascribed to the behavior of
cells in a higher-level description; e.g., the
expression of the gene GATA3 in CD4 T
lymphocytes in a certain experimental condition
gives indication about the differentiation state of
these cells, on the pattern of cytokine secreted and
ultimately on the type of the immune response
(Santoni et al., 2008); this is an information that is
relevant to the construction of a computational
model of the immune response.
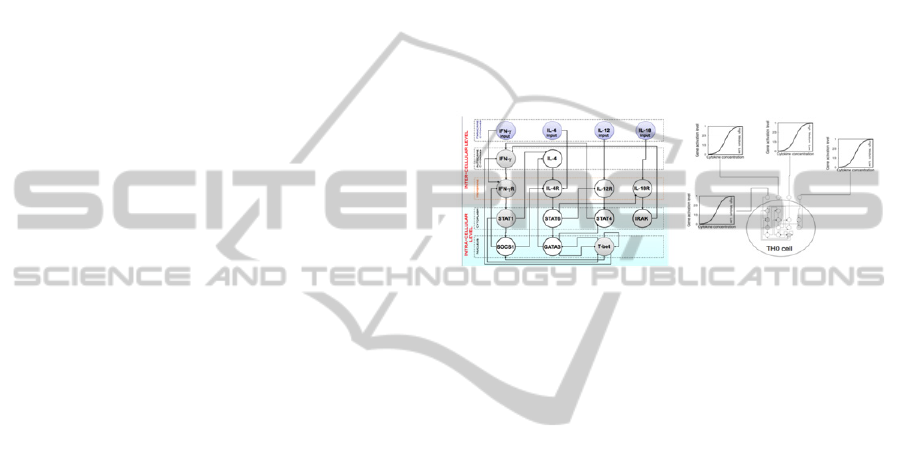
For example, we have implemented a gene
regulatory network (GRN) of the intracellular-level
gene expression dynamics to characterize the
Th1/Th2 cell differentiation, a phenomena that takes
place at the cellular (mesoscopic) level. The GRN
used represents the most extensive attempt to model
the regulatory network controlling the differentiation
of TH lymphocytes to date (Mendoza, 2006). Before
integrating the minimalistic Boolean network
dynamics in an agent-based model of the cell-cell
interaction, we identified the genes coding for
membrane receptors and those coding for soluble
molecules to be secreted by the cell, with the idea of
interpreting the former as the “input” and the latter
as the “output” of the cell (left panel of Figure 2).
Then we analysed the network Boolean dynamics
using classical logical methods to identify the
asymptotic regimens. In particular, three ‘attractors’
with relevant biological meaning were identified:
two leading to TH1 (P1 and P2) and one to TH2 (P3)
phenotype. For each time step of the simulation each
undifferentiated T helper cell would individually
transduce the input signals coming from the
extracellular space through the cell receptors (right
panel of Figure 2) into a micro-dynamics of the gene
regulatory network eventually falling (or not) in one
of the attractors. In the case one of the possible
attractors is reached, then rule is fired and the cell
becomes a Th1 or Th2, otherwise the cell remains in
the undifferentiated state. More formally, we
obtained a partition of the space of all possible
configurations Ω0,1
(17 are the genes of the
GRN) considering hyper spheres of radius two
centred in P1, P2 and P3, that is,
∈
Ω: , 2, 1,2,3, where ∀, ∈
Ω,
,
∑
and
Ω
∪
∪
) is the remaining space. Note that
∩
∅ while
∪
∩
∅. The
rule states that, at time 1, undifferentiated Th
cells at time , whose internal network state belongs
to
∪
, are marked as Th1; those with internal
state in
are marked Th2 and all the rest do not
differentiate.
Figure 1: Left panel: The GRN used to control the
differentiation of Th cells. Nodes correspond to
genes/molecules involved in the Th1/2 switch. Connectors
ending with an arrow indicate activation while those
ending with a dot indicate inhibition. Right panel: The
differentiation of each uncommitted Th cell depends on
the concentration of input cytokines surrounding it. These
cytokines determine the activation level of the
corresponding input nodes, i.e., if cx denotes the
concentration of an input cytokine, then the activation
level of the corresponding input node is given by [m ·
c
2
/(θ
2
+ c
2
)] where [x] denotes the smallest integer greater
than x, m are the activation levels and θ is a constant.
The resulting automaton was able to reproduce a
dynamics that was consistent with macroscopic
observable phenomena at the cell population level
still remaining compatible with a realistic gene
expression profile at the microscopic level (Santoni
et al., 2008). This example shows that the two levels
of description (intracellular and extracellular) can
realistically be integrated supposed that (i) the
intracellular gene regulatory network is biologically
sound and allows for relevant asymptotic regimens
and (ii) the stable dynamics at the lower level can be
rationally translated into an action (the rule) at the
upper level.
As already mentioned, mathematical models that
try to describe such mechanisms, usually fix a
spatial and the temporal scale and describe the
system with a mathematical or computational (i.e.,
algorithmic) formalism (Dada and Mendes, 2011;
Engler et al., 2009; Qu et al., 2011). Computers do
ComputationalBiologyModelingacrossDifferentScales
619

the rest as they provide the dynamics by executing
(resolving) the rules just described in mathematical
formalism. The dynamics is dependent on
parameters and initial conditions so that one
generally tries hypothetical scenarios modifying
those initial conditions to get a feeling of the
systems behavior (Meier-Schellersheim et al., 2009;
Schenell, 2007). This process leads itself in
discovering new knowledge. However, the problem
is that the real system is in general not isolated
hence a local description is not sufficient to disclose
crucial mechanisms. It comes quite clear that one of
the reasons why biological phenomena are
intrinsically complex is because they are influenced
by variables that are outside a single level of
space/temporal description.
If we take into consideration the space, a good
way to define a scale is to selectively assign
processes to their position within a biological
hierarchy i.e., whether they represent interactions
between organs, within a tissue, between cells, and
so on. We can refer to these hierarchical positions as
levels of a biological organization. A relevant note
to this question is expressed in (Southern et al.,
2008), namely, when comparing different organisms
with each other, the specific spatial-temporal scales
in standard international units may be quite
different, even when looking at the same level of
biological organization and it would therefore be
beneficial for multi-scale modeling in bio-medicine
to refer to these levels of organization.
Biological systems can be thought as hierarchical
structures, i.e., genes that encode proteins; proteins
that are building blocks of organelles and cells; cells
that form tissue and organ; organs that form
organisms; and organisms that give origin to
individuals and populations. Different levels
communicate each other in the sense that lower
levels affect the higher ones and vice versa. For
example proteins regulate gene expression.
Therefore, in a biological system, interactions can
occur both at the same scale (such as interactions
between different cells) as well as between scales.
This originates a very complex system in which one
has to deal with multiple spatial and temporal scales
and feedback loops.
In theory, one can develop a model of a
biological system (such as a cardiac cell or the heart)
consisting of the genes and proteins, or even the
atoms. In practice however, existing computational
tools are yet insufficient for this task. It should be
noted that experiments are done at many scales,
ranging from single molecules or proteins to whole
organs and organisms, and therefore, experimental
information exists at different scales. Therefore,
relying on different experimental data, a model can
be formulated using two main approaches, i.e., top-
down or bottom-up (Alberghina and Westerhoof,
2008).
If one chooses to take into account the individual
elements and their interactions, studying the
resulting biological system as a consequence of the
emergent behavior of its single components, then the
bottom-up approach takes place. The advantage of
this type of approach is that it is adaptive and robust,
in the sense that if the available biological
knowledge varies, one can adapt the new knowledge
to the specific components of the model, in a very
selective way. Moreover this kind of approach is
suitable for studying the emergent properties of
systems consisting of a large number of interacting
elements. The intensive computer power required is
the main disadvantage for the bottom-up approach
and can be sometimes even prohibitive. Moreover,
the model itself can become too complicated to
control.
Instead, one can decide not to look straight into
the details of the individual elements, but to consider
the system at the macroscopic level, using
experimental observations as guidelines during the
formulation of the model. The clear advantage of
this approach is that it is relatively simple. On the
other hand, the adaptability and the robustness of the
model are less evident compared with the bottom-up
approach. Moreover, it should be highlighted that
the variables and parameters in these models are
largely phenomenological without direct connection
with detailed physiological parameters. Due to this
reason, it may sometimes happen that the top-down
approach does not correctly reveal the actual
responsible mechanism, e.g., when there are
multiple mechanisms for the same behavior or a
single mechanism resulting in multiple phenomena.
When existing components have to be integrated
with some new part a third design principle, named
“middle-out”, is used (Hunter and Viceconti, 2009).
3 MODELING ACROSS
DIFFERENT
SCALES – FILLING THE GAP
Going from the lowest scale to higher levels one can
choose among different modeling choices. Intra-
cellular modeling approaches aim at a detailed,
mechanistic description of molecular processes
occurring inside single cells. These models usually
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
620

adopt the differential equation description to predict
the molecular kinetics of specific cellular pathways
starting from experimentally determined parameters.
These models consist of mass action or Michaelis-
Menten kinetic rate-law equations describing the
changes of molecular concentrations. An example of
a bi-domain model describing a phenomenon at a
level that originates from the microscopic dynamics
at a smaller space scale is the wave propagation in
reactive media belonging to the class of the so-called
Belousov– Zhabotinsky reaction. In a simple form
(called the “oregonator” model) it may be
understood in terms of the following schema (Tyson,
1994) including an autocatalytic reaction
A+Y→X+P, X + Y → 2P, A + X → 2X + 2Z, 2X
→ A + P,
B + Z → hY + Q, where the variables
represents concentrations of specific molecules (e.g.,
bromomalonicacid, carbon dioxide, etc.) and h is a
constant. Translated to ordinary differential
equations the system is dX/dt =AY −XY +AX−2X
2
,
dY/dt = AY − XY + hBZ, dZ/dt = 2AX – BZ, where
A, B and P are held constant. The solution of this
system has an oscillatory dynamics that, transposed
to two spatial dimensions, describes a propagating
wave. The bi-domain “nature” of the model in this
example lays in the emergence of the wave at a level
that is above the one chosen to describe the
phenomena, that is the molecular level of the
reactants (Murray, 2003).
An alternative to differential equations for intra-
cellular models is the microsimulation of reactions
within cells where the number of reagents is a small
number (due to current computational limitations).
The method developed many decades ago and
known as the Gillespie algorithm (Gillespie, 1976;
Gillespie, 1977) allows to accurately simulating
chemical or biochemical systems of reactions
generating a statistically correct trajectories as
possible solutions of a stochastic equation as for
example the differential equations corresponding to
the time-evolution of stochastic processes that
proceed by jumps (e.g., Markov jump process
(Bailey, 1990)). A simplified version of this
equation is the master equation describing the time
evolution of the probability
P
of a system to be in a
set of states with regard to a continuous time
variable t. The most familiar form of a master
equation is a matrix form
dP
M
P
dt
(1)
where M is the matrix specifying the connections. At
a higher scale level of description, tissues or whole
organs are modeled in two different ways: either as
functional compartments or system units or as a
collection of microscopic components (e.g., cells).
These two modeling paradigm use a completely
different point of view in describing a functional
unit as a tissue or organ. In the former case the organ
is seen as a black box with known input-output
relationship. This relation is typically derived from
known facts and ultimately realized by differential
equations linking stimulus with response or input to
output or causes to effects. These kind of
phenomenological models do not attempt to give an
explanation of the observed behavior whereas they
aim merely at reproducing it. They are quite useful
when combined together to offer a bigger picture.
The latter modeling paradigm proposes to represent
a tissue as an array of individual units (i.e., cells)
exchanging signals with the environment. A
noticeable example of these multicellular systems
has been originally developed to study the growth of
solid tumors (Drasdo et al., 1995; Drasdo, 2000),
and has later on been applied to simulate the
function (the regeneration) of complex organs like
the liver (Hoehme et al., 2010).
There are a number of ongoing projects whose
aim is to simulate a whole cell (e.g., virtual cell
(Schaff et al., 1977), e-cell (Normile, 1999;
Takahashi et al., 2004)), whereas efforts aiming at
simulating whole systems or organs are, for
example, models of the heart (Hunter and Nielsen,
2005), of the liver (Holzhütter et al., 2012), and of
the skeletal system (Viceconti, 2012). Other efforts
aim at creating computational platforms suite to
integrating various physiological processes (Eissing
et al., 2011). These are integrative systems biology
challenges that target the simulation of complex
biological systems through multi scale integration of
different mathematical and computational models.
The approach is the so-called middle-out strategy
proposed by Brenner, (1998) and Noble (2002;
2006), based on the principle that, in biology, there
is no privileged level for the description of a certain
phenomenon and that the inter-level causal
relationships are driven by interactions between
multiple levels. An application of the same modeling
principle to nutritional sciences can be found in de
Graaf et al., (2009) where the authors describe how
multi-scale models integrating processes from the
cellular up to the physiological levels are indeed
necessary in answering important nutritional
questions.
The use of different modeling paradigms
however, introduces gaps between scales. Multi-
scale modeling, besides modeling the individual
system components, needs to address the issue of
how to bridge the gaps between different
ComputationalBiologyModelingacrossDifferentScales
621

methodologies and between models at different
scales. Unfortunately, there is not a specific or
simple way to achieve this goal, but there are quite a
number of empirical principles and methods that can
provide some hint. For instance, adaptive mesh
refinement in lattice models (Plewa et al., 2005) is
used to scale down the details of a certain process,
the Hidden Markov Models (Baum and Petrie, 1966)
are used to deduce higher scale logics from the
observation of lower scale patterns, equation free
methods (Kevrekidis et al., 2003) based on the
execution of microscopic simulation models
allowing for computing the evolution equation of a
system at a higher (e.g., coarse) level, etc.
Systems biology is the main area in which one
can find this help. The goal of systems biology is to
consider a biological system from a holistic
perspective, and use both experiments and modeling
and the interactions between experiments and
modeling to reveal how the system behaves (Kitano,
2002; Kohl et al., 2010).
Specific modeling choices at a lower length scale
favor the integration of information at higher scales
and vice versa. For example, the individual- or
agent-based modeling approach at the mesoscopic
level (Castiglione et al., 2007) can be integrated to
the microscopic intracellular description for which
we can adopt either the continuous approach (as in
Ribba et al., 2006, that integrates cell cycle
regulation and macroscopic tumor dynamics with
the aim with the aim of mathematically investigating
this therapeutic failure the anti-metastatic agents
called inhibitors of metalloproteinases), or Boolean
networks to model intracellular events (like the
regulation of gene activation as in the differentiation
of T lymphocytes (Santoni et al., 2008)). In other
words, taken out the necessary approximation, a
fruitful approach in constructing large-scale
mechanistic models is given by combining
mechanistically detailed kinetic models (either
continuous - equations based - or discrete - boolean
networks) and coarse-grained (i.e., individual- or
agent-based) models (Smallbone et al., 2007).
Interestingly, it has been shown recently that
complex system behavior is often largely defined by
the interaction topology among the various model
components (Brown et al., 2004; Gutenkunst et al.,
2007). This finding further supports the expectation
that in order to obtain meaningful predictions most
likely only a few molecular processes need to be
described in great detail with precise parameters
estimates, while the rest of the system can be
described using the coarse-grained interaction
topology (de Graaf et al., 2009).
The very multi-scale nature of novel models in
computational biology makes their development
particularly challenging, not just from a biological
point of view but also from a mathematical and
computational perspective. Moreover, given the
availability of already published models targeting a
single scale, the sharing and reusing of such models
has become an issue. A prominent attempt at solving
this problem is provided by the Physiome Project
(Bradley et al., 2011; Hunter and Borg, 2003), which
aims at developing a framework for the modeling of
the “whole” human body. As part of that initiative,
the mark-up language CellML was introduced with
the aim of establishing a world-wide-adopted
standard in the development of cellular level that are
modeled as sets of ODEs (Garny et al., 2008).
Similarly, FieldML has been defined to model
processes on the tissue and organ level that are
represented as sets of PDEs (Christie et al., 2009).
Along with CellML, another standard called
Systems Biology Markup Language (SBML) (Hucka
et al., 2003) has been proposed and is now beginning
to make a significant impact on the modeling
community as a means to exchange models.
However, neither CellML nor SBML include
explicit directives to deal with the problem of
implementing a multi-scale computational model,
although there are attempts to address this issue
(Baylei, 1990).
Regardless the integration framework one
decides to use there are few aspects that need to be
taken into account when developing a multi-scale
model. In general, the time scales on which the
lower-level processes occur are much faster than
those on which the higher-level processes occur.
Usually this means that the lower-level processes
can be assumed to occur instantaneously and can
therefore be included as a representation of some
kind of field at the higher level. The switch to a
model at a higher level of organization is usually
determined by the need to ensure that the
calculations can be performed in reasonable time
(Southern et al, 2008). When coupling together
independent models of processes that occur on
different scales or as part of different physical
systems (as is in multi-organ systems) it is enticing
to simply couple existing components (i.e., software)
for the separate models to one another. However,
this does not take into account how inaccuracies in
the values of the variables that are passed between
the two models may affect the combined model -
one variable may be accurate enough in one model
but when these models are coupled may first
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
622

introduce errors into the solution of the other model,
and in turn the solution of the combined model. In
order to prevent these inaccuracies from occurring
one should consider the whole as a single model
rather than the combination of two simpler ones. For
instance, we can consider a microscopic simulator at
the cellular level can be coupled with the description
of the intracellular signaling activating a specific
cellular pathway. In this example the differentiation
of T lymphocytes into the phenotypes Th1, Th2,
Treg and Th17 is described at a cellular level by
means of individual entities (e.g., agent-based)
whereas the gene regulation is described (at variance
with the example above which use a Boolean
network) by a system of differential equations
describing activation level of each gene of the gene
network represented with the following equation
()
()
(1 )(1 )
i
i
hC
Ch
i
ii
hC
Ch
dx
ee
x
dt
ee
(2)
where
is the activation level of the i
th
gene,
and
are parameters relative to the network topology
and C is a constant (Mendoza and Pardo, 2010).
Here the lower level description of gene activation is
determined at each upper-level time step by solving
the system of ODEs, and the cell differentiation is
executed at the upper level on the basis of the
information coming from the gene expression levels.
This procedure is iteratively executed at each time
step and for each lymphocyte.
4 CONCLUSIONS
In the study of complex biological phenomena it is
necessary to develop a unified view of the various
mechanisms in action and of the causal relationships
among different parts of that complex system (Di
Ventura et al., 2006; Kitano, 2002). In this article we
have briefly described the problems faced when one
wants to link mathematical or computational models
across different time and length scales.
In many areas of biology and physiology, multi-
scale and multi-physics models are very much
acclaimed, Although there exist an abundant
literature for multi-scale models in science and
engineering domains (Fish, 2010; Weinan, 2011), a
lot remains to be done in terms of translating these
mathematical theories and methodologies to the
domains of biology and physiology (Evans et al.,
2008; Caiazzo et al., 2011; Tahir et al., 2011).
A key unsolved issue is how to represent
appropriately the dynamical behaviors of a high-
dimensional model of a lower scale by a low-
dimensional model of a higher scale, so that it can be
used to investigate complex dynamical behaviors at
even higher scales of integration (Qu et al., 2011).
The use of different modeling techniques,
introduces gaps between scales. Multi-scale
modeling, besides modeling the system, needs to
address the issue of how to bridge the gaps between
different methodologies and between models at
different scales. Unfortunately, there is no specific
or simple way to tell how to achieve this objective,
but there are empirical principles and methods that
can be of help. The goal of computational systems
biology to consider a biological system from a
holistic perspective, and use both experiments and
modeling to reveal how the system behaves (Kitano,
2002; Kohl et al., 2010). It is certainly one of the
main research fields that can benefit from the use of
multi-scale models and, at the same time, provide
methodologies for their development.
ACKNOWLEDGEMENTS
This work was partially supported under the EC
contract FP7-ICT-2011-9, No.600803 (MISSION-
T2D)".
REFERENCES
Alberghina L, Westerhoof HV. Systems Biology.
Definitions and perspectives. Heidelberg; Springer,
2008.
Bailey NTJ. The Elements of Stochastic Processes with
Applications to the Natural Sciences. New York;
Wiley, 1990.
Bassingthwaighte JB, Chizeck HJ, Atlas LE, et al.
Multiscale modeling of cardiac cellular energetics.
Ann. N. Y. Acad. Sci. 2005; 1047:395-424.
Baum LE, Petrie T. Statistical Inference for Probabilistic
Functions of Finite State Markov Chains. The Annals
of Mathematical Statistics 1966; 37(6):1554-1563.
Bradley C, Bowery A, Britten R, et al. OpenCMISS: a
multi-physics & multi-scale computational
infrastructure for the VPH/Physiome project. Prog.
Biophys. Mol. Biol. 2011; 101:32-47.
Brenner S. Biological computation. In: Bock G, Goode J
(Eds.). The limits of reductionism in biology. Novartis
Foundation Symposium, vol. 213. London UK: Wiley,
1998, 106-116.
Brown K. S., Hill C. C., Calero G. A., et al. The statistical
mechanics of complex signaling networks: nerve
growth factor signaling. Phys. Biol. 2004; 1(3-4):184-
195.
Caiazzo A, Evans D, Falcone J-L, et al. A Complex
Automata approach for in-stent restenosis: Two-
ComputationalBiologyModelingacrossDifferentScales
623

dimensional multiscale modelling and simulations. J.
Comp. Science, 2011; 2(1):9-17.
Castiglione F, Liso A, Bernaschi M, Succi S. Microscopic
simulation in biology and medicine. Current Medicinal
Chemistry 2007; 14(6):625-637.
Christie GR, Nielsen PMF, Blackett SA, et al. FieldML:
concepts and implementation. Phil. Trans. R. Soc. A
2009; 367(1895):1869-1884.
Coveney PV, Fowler PW. Modelling biological
complexity: a physical scientist’s perspective. J. R
Soc. Interface 2005; 2:267-280.
Dada JO, Mendes P. Multi-scale modelling and simulation
in systems biology. Integr. Biol. 2011; 3:86-96.
Deane C. M., Salwinski L, Xenarios I, et al. Protein
interactions: two methods for assessment of the
reliability of high throughput observations. Mol. Cell.
Proteomics 2002; 1:349-356.
de Graaf A. A., Freidig AP, De Roos B, et al. Nutritional
Systems Biology Modeling: From Molecular
Mechanisms to Physiology. PLoS Comp. Biol.; 2009
5(11):e1000554.
Di Ventura B, Lemerle C, Michalodimitrakis K, et al.
From in vivo to in silico biology and back. Nature
2006; 443(7111):527-533.
Drasdo D, Kree R, McCaskill JS. A Monte Carlo Model to
tissue cell populations. Phys. Rev. E 1995;
52(6):6635-6657.
Drasdo D. Buckling Instabilities in One-Layered Growing
Tissues. Phys. Rev. Lett.; 2000 84:4424-4427.
Evans D, Lawford P, Gunn J, et al. The application of
multiscale modelling to the process of development
and prevention of stenosis in a stented coronary artery.
Phil.Trans. R. Soc. A, 2008; 366:3343-3360.
Eissing T, Kuepfer L, Becker C, et al. A computational
systems biology software platform for multiscale
modeling and simulation: integrating whole-body
physiology, disease biology, and molecular reaction
networks. Frontiers in Physiology; 2011 2(4):1-10.
Engler AJ, Humbert PO, Wehrle-Haller B, et al.
Multiscale modeling of form and function. Science
2009; 324:208-212.
Falcone J, Chopard B, Hoekstra A. MML: towards a
Multiscale Modeling Language. Procedia Computer
Science, 2010; 1(1):819-826.
Fish J (Ed.) Multiscale Methods, bridging the scales in
science and engineering. Oxford: Oxford University
Press, 2010.
Garny A, Nickerson DP, Cooper J, et al. CellML and
associated tools and techniques. Philos Transact A
Math Phys Eng Sci. 2008; 366(1878):3017-3043.
Gillespie DT. A General Method for Numerically
Simulating the Stochastic Time Evolution of Coupled
Chemical Reactions. J. Comp. Phys. 1976; 22 (4):403-
434.
Gillespie DT. Exact Stochastic Simulation of Coupled
Chemical Reactions. J. Phys. Chem. 1977;
81(25):2340-2361.
Grima R. Multiscale modeling of biological pattern
formation. Curr. Top. Dev. Biol. 2008; 81:435-460.
Gutenkunst RN, Waterfall JJ, Casey FP, et al. Universally
sloppy parameter sensitivities in systems biology
models. PLoS Comput. Biol. 2007; 3:1871-1878.
Hoehme S, Brulport M, Bauer A, et al. Cell alignment
along micro-vessels as order principle to restore tissue
architecture during liver regeneration: from
experiment to virtual tissues and back. Proc. Natl.
Acad. Sci. USA; 2010 107(23):10371-10376.
Holzhütter H-G, Drasdo D, Preusser T, et al. The virtual
liver: a multidisciplinary, multilevel challenge for
systems biology. Wiley Interdiscip. Rev. Syst. Biol.
Med. 2012; 4(3):221-235.
Hucka M, Finney A, Sauro HM, et al. The Systems
Biology Markup Language (SBML): A Medium for
Representation and Exchange of Biochemical Network
Models. Bioinformatics 2003; 9(4):524-531.
Hunter PJ, Borg TK. Integration from proteins to organs:
the Physiome Project. Nat. Rev. 2003; 4:237-243.
Hunter P., Nielsen P. A., Strategy for integrative Compu-
tational physiology. Physiology 2005; 20:316-325.
Hunter PJ, Viceconti M. The VPH-Physiome Project:
Standards and Tools for Multiscale Modeling in
Clinical Applications. IEEE Reviews in Biomedical
Engineering 2009; 2:40-53.
Joshi H, Singharoy AB, Sereda YV, et al. Multiscale
simulation of microbe structure and dynamics. Prog.
Biophys. Mol. Biol. 2011; 107:200-217.
Kevrekidis IG, Gear CW, Hyman JM, et al. Equation-free,
coarse-grained multiscale computation: enabling
microscopic simulators to perform system-level tasks.
Comm. Math. Sciences 2003; 1(4):715-762.
Kitano H. Systems Biology: A Brief Overview. Science
2002; 295(5560):1662-1664.
Kohl P, Crampin EJ, Quinn TA, et al. Systems biology: an
approach. Clin. Pharmacol. Ther.; 2010 88:25-33.
Meier-Schellersheim M., Fraser I. D., Klauschen F.,
Multiscale modeling for biologists. Wiley Interdiscip.
Rev. Syst. Biol. Med. 2009; 1:4-14.
Mendoza L. A network model for the control of the
differentiation process in Th cells. Bio. Systems 2006;
84:101–114.
Mendoza L, Pardo F. A robust model to describe the
differentiation of T-helper cells. Theory Biosci. 2010;
129(4):283-293.
Murray JD. Mathematical Biology vol I and vol II. NY:
Springer-Verlag, 2003.
Murtola T, Bunker A, Vattulainen I, et al. Multiscale
modeling of emergent materials: biological and soft
matter. Phys. Chem. Chem. Phys. 2009; 11:1869-
1892.
Noble D. Modeling the heart – from genes to cells to the
whole organ. Science 2002; 295:1678-1682.
Noble D. The Music of Life. Biology Beyond the
Genome. Oxford UK: Oxford University Press, 2006.
Normile D. Building Working Cells ’in Silico’. Science
1999; 284(5411):80-81.
Plewa T, Linde T, Weirs VG. Adaptive Mesh Refinement
- Theory and Applications. Lect. Notes Comp. Sci.
and Eng. 2005; 41:341-350.
Qu Z, Garfinkel A, Weiss JN, Nivala M. Multi-scale
modeling in biology: How to bridge the gaps between
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
624

scales? Prog. Biophys. Mol. Biol. 2011; 107:21-31.
Ribba B, Sautb O, Colin T, et al. A multiscale
mathematical model of avascular tumor growth to
investigate the therapeutic benefit of anti-invasive
agents. J. Theo. Biol. 2006; 243(4):532-541.
Santoni D, Pedicini M, Castiglione F. Implementation of a
regulatory gene network to simulate the TH1/2
differentiation in an agent-based model of hyper-
sensitivity reactions. Bioinformatics 2008; 24:1374-
1380.
Schaff J, Fink CC, Slepchenko B, et al. A general
computational framework for modeling cellular
structure and function. Biophys. J. 1997; 73:1135-
1146.
Schnell S, Grima R, Maini PK. Multiscale modeling in
biology. Am. Scientist 2007; 95:134-142.
Smallbone K, Simeonidis E, Broomhead DS, et al.
Something from nothing: bridging the gap between
constraint-based and kinetic modelling. FEBS J 2007;
274:5576-5585.
Sloot PMA, Hoekstra AG. Multi-scale modelling in
computational biomedicine. Brief. Bioinform. 2010;
11(1):142-152.
Southern J, Pitt-Francis J, Whiteley J, et al. Multi-scale
computational modelling in biology and physiology.
Prog. Biophys. Mol. Bio. 2008; 96(1-3):60-89.
Tahir H, Hoekstra AG, Lorenz E, et al. Multi-scale
simulations of the dynamics of in-stent restenosis:
impact of stent deployment and design. Interface
Focus 2011; 1(3):365-373.
Takahashi K, Kaizu K, Hu B, et al. A multi-algorithm,
multi-timescale method for cell simulation.
Bioinformatics 2004; 20:538-546.
Tyson JT. What Everyone Should Know about the
Belousov-Zhabotinsky Reaction. In: Levin SA (Ed.).
Frontiers in Mathematical Biology. NY: Springer
Verlag, 1994, 569–587.
Viceconti M. Multiscale Modeling of the Skeletal System.
NY: Cambridge Univ. Press, 2012.
Weinan E. Principles of multiscale modeling. NY:
Cambridge Univ. Press, 2011.
ComputationalBiologyModelingacrossDifferentScales
625
