
Cluster Detection and Field-of-View Quality Rating
Applied to Automated Pap-smear Analysis
Marine Astruc
1
, Patrik Malm
2
, Rajesh Kumar
3
and Ewert Bengtsson
2
1
Ecole Centrale Nantes, Nantes, France
2
Centre for Image Analysis, Division of Visual Information and Interaction, Department of Information Technology,
Uppsala University, Uppsala, Sweden
3
Centre for Development of Advanced Computing, Thiruvananthapuram, India
Keywords:
Pap-smear, Automated Screening, Cluster Detection, Field-of-View Analysis.
Abstract:
Automated cervical cancer screening systems require high resolution analysis of a large number of epithelial
cells, involving complex algorithms, mainly analysing the shape and texture of cell nuclei. This can be a very
time consuming process. An initial selection of relevant fields-of-view in low resolution images could limit
the number of fields to be further analysed at a high resolution. In particular, the detection of cell clusters
is of interest for nuclei segmentation improvement, and for diagnostic purpose, malignant and endometrial
cells being more prone to stick together in clusters than other cells. In this paper, we propose methods aiming
at evaluating the quality of fields-of-view in bright-field microscope images of cervical cells. The approach
consists of the construction of neighbourhood graphs using the nuclei as the set of vertices. Transformations
are then applied to such graphs in order to highlight the main structures in the image. The methods result in
the delineation of regions with varying cell density and the identification of cell clusters. Clustering methods
are evaluated using a dataset of manually delineated clusters and compared to a related work.
1 INTRODUCTION
According to the World Health Organization (WHO)
cervical cancer is the second most common type
of cancer among women, annually killing close to
300,000 world wide. Out of these deaths 86% oc-
cur in developing countries (WHO/ICO Information
Centre on HPV and Cervical Cancer (HPV Informa-
tion Centre), 2012). An important reason to the large
difference is the absence of organized screening pro-
grammes using the Papanicolaou test (Pap test) de-
veloped by Dr. Georges Papanicolaou in the 1940s
(Papanicolaou et al., 1943). For this test cells are ob-
tained from the uterine cervix through a simple scrap-
ing/brushing operation and smeared onto a glass slide
of 25x75 mm, fixated and stained and then inspected
through a normal light microscope at high resolution.
Based on a Pap-test, trained cytologists can not
only find proof of invasive cancer but also detect cer-
tain cancer precursors, allowing for early and effec-
tive treatment. If detected early, invasive cancer is
curable and the 5-year survival rate as high as 92%
(Saslow et al., 2012).
Most screening programmes revolve around vi-
sual screening performed by cytotechnicians in spe-
cialized laboratories. The screening work is tedious
and, often due to fatigue, error prone. Because of the
hazards of fatigue some recommendations say that a
cytotechnician should not work more than 7 hours a
day and analyse no more than 70 samples (Elsheikh
et al., 2012). This implies an average time to analyze
a specimen of 6 minutes, a very short time given the
complexity of the task.
Although the Pap test has shown its worth through
decades of use it is hampered by a number of difficul-
ties, e.g., variable smear thickness, uneven cell distri-
bution, dense cell groupings (clusters), obscuring ele-
ments such as blood and inflammatory cells and vari-
able fixation and staining results (Grohs and Husain,
1994).
To overcome some of the human limitations sev-
eral attempts to automate the screening process have
been made since the 1950s with varying degree of
success. Today there are systems that are able to per-
form a scan of a sample but they all have in common
that they require very specific sample preparation and
are complicated and expensive to run (Bengtsson,
2005).
355
Astruc M., Malm P., Kumar R. and Bengtsson E. (2013).
Cluster Detection and Field-of-View Quality Rating - Applied to Automated Pap-smear Analysis.
In Proceedings of the 2nd International Conference on Pattern Recognition Applications and Methods, pages 355-364
DOI: 10.5220/0004254803550364
Copyright
c
SciTePress

A fundamental problem in developing a screening
system is the vast areas that need to be analysed. A
regular PAP-smear covers an area of at least 25x50
mm. The resolution needed for determining the ma-
lignancy of a cell leads to a pixel size of around 0,2
microns. This translates to 31 billion pixels on a spec-
imen. Handling this amount of data in a few minutes
poses serious challenges both on the initial scanning
side and on the subsequent data analysis side. One
way of improving the situation is to use a modified
technique for depositing the slides on the specimen.
So called Liquid Based Preparations, LBP, typically
deposit the material in a circle with a diameter of 20
mm. This reduces the number of pixels to around 8
billion, still a substantial number, at the cost of a sub-
stantially more complex and costly slide preparation
procedure. For the final analysis of a cell to be reliable
it has to be in perfect focus and the algorithms to ex-
tract the relevant features are typically quite elaborate
and thus time consuming. Autofocus and complex
analysis algorithms thus make the automated screen-
ing problem even more challenging.
One way to attack this difficulty is to have a two
stage approach, an initial search phase for areas or
cells of interest followed by a detailed analysis of
the interesting regions. This approach was first sug-
gested and analyzed by Poulsen (Poulsen, 1973) and
later implemented in the Diascan system (Nordin,
1989). There have been huge improvements in scan-
ning and computer technology since the 1970-80 ies
when these projects were conducted but the funda-
mental problem holds. We thus need to find efficient
ways of determining where on the slide we should fo-
cus our attention to reach a reliable decision about
whether the specimen is normal or possibly show
some abnormalities.
The initial analysis can be conducted of fields of
view of lower resolution and with less stringent re-
quirement on perfect focus. Whether these fields are
obtained by merging pixels or subsampling images
scanned at full resolution or by a separate scan of
the specimen with different optics is a technical is-
sue that requires a complicated technical/economical
analysis to find the best solution for a particular set-
ting. We will not discuss those issues further in this
paper. For the study in this paper we have worked
with images with a pixel size of 0.5 microns and with
a single rough focus setting. This represents between
one and two orders of magnitude less data than the
perfectly focused, high resolution images needed for
the final analysis.
The task of this low resolution analysis is to find
areas that should be analysed more in detail. This
will trivially mean to discard completely empty ar-
eas or areas where the cells are spread so dense that
they cannot be resolved. We will be looking for areas
with suitable density of cells of potential diagnostic
interest. This could be extended to only look for cells
that are larger than normal, since malignant cells usu-
ally are larger than normal ones. But stretching this
criterion too far risks leading to missing some spec-
imens where the malignant cells are of normal size
(such malignancies exists). So we will be counting
cells that are of a relevant size for further analysis,
not only cells significantly bigger than normal.
Another important task is to look for clusters. It is
known that malignant cells tend to cluster more than
normal ones so when the human screener see a clus-
ter of cells they take an extra look. We should thus
note and flag the appearance of clusters in the anal-
ysed fields.
So to summarize we will in this paper present
a study of image fields of moderate resolution from
standard PAP smears and LBP specimens generating
data that can be used to prioritize which areas should
be used for the subsequent more expensive high reso-
lution analysis. Thus optimizing the overall through-
put of a system without sacrificing detection qual-
ity. The methods described in this paper can also
work towards the overall classification task by locat-
ing diagnostically important structures that are often
overlooked in conventional cell by cell classification
schemes. We have not found any studies in the re-
cent literature with this goal, most papers on PAP-
smear analysis deal with segmentation or classifica-
tion problems of images at a single resolution level.
However, Raymond et al. (Raymond et al., 1993)
made use of graphs and mathematical morphology
to analyse neighbourhood relationship between cells
in the study of germinal centers. Also, in a recent
publication Chandran et. al (Chandran et al., 2012)
presented a method for detecting clusters in cervical
smears that is of interest and that is used for compari-
son in this paper.
2 MATERIALS AND METHODS
2.1 Microscope Setup
The images were acquired using an Olympus BX51
optical microscope with a 20X, 0.75 NA objective and
a Hamamatsu ORCA 05G monochrome digital cam-
era providing images of 1344 x 1024 with an effective
square pixel size of 0.5 microns. The illumination was
filtered through a narrow green filter centered at 570
nm in order to optimize nuclear contrast.
ICPRAM2013-InternationalConferenceonPatternRecognitionApplicationsandMethods
356
2.2 Methodology
The methods developed to analyse an image field
from a cervical smear sample are based on transfor-
mations applied to graphs that are built using the cell
nuclei as vertices. The first step consists of the seg-
mentation of the cell nuclei. Then, neighbourhood
graphs are constructed and several transformations
are applied to the different graphs in order to separate
the image field into three regions according to how
densely the nuclei are spaced and to locate the cells
belonging to clusters.
2.3 Nuclei Segmentation
In order to build graphs on an image field, we need to
segment the cell nuclei. A preprocessing step is used
to reduce the background noise and improve the im-
age quality. It consists of the implementation of a me-
dian filter. The contrast of the grayscale image is en-
hanced using Contrast-Limited Adaptive Histogram
Equalization (Zuiderveld, 1994). As a first stage of
the segmentation, grayscale morphological closings
with annular flat structuring elements are applied to
identify nuclei-like objects, which will serve as seeds.
The nuclear boundaries are then delineated using
seeded watershed segmentation (Moshavegh et al.,
2012).
The resulting segmented nuclei-like objects are
then classified into two groups, nuclei and artifacts,
using Support Vector Machine. To evaluate the classi-
fication results, a Graphical User Interface (GUI) was
developed to permit a user to identify nuclei in a set of
images. A total of 24 images, belonging to different
samples, have been marked. 2478 objects were man-
ually identified as nuclei and 5474 identified as arti-
facts. The sensitivity, the specificity and the accuracy
of the classification are 96,6%, 86,0% and 89,3% re-
spectively. Although the results of segmentation and
classification present some errors (missing nuclei, re-
maining artifacts), the objects identified as nuclei af-
ter the classification will be considered as true nuclei
throughout the analysis. These imperfections affect
the results obtained by the methods herein presented.
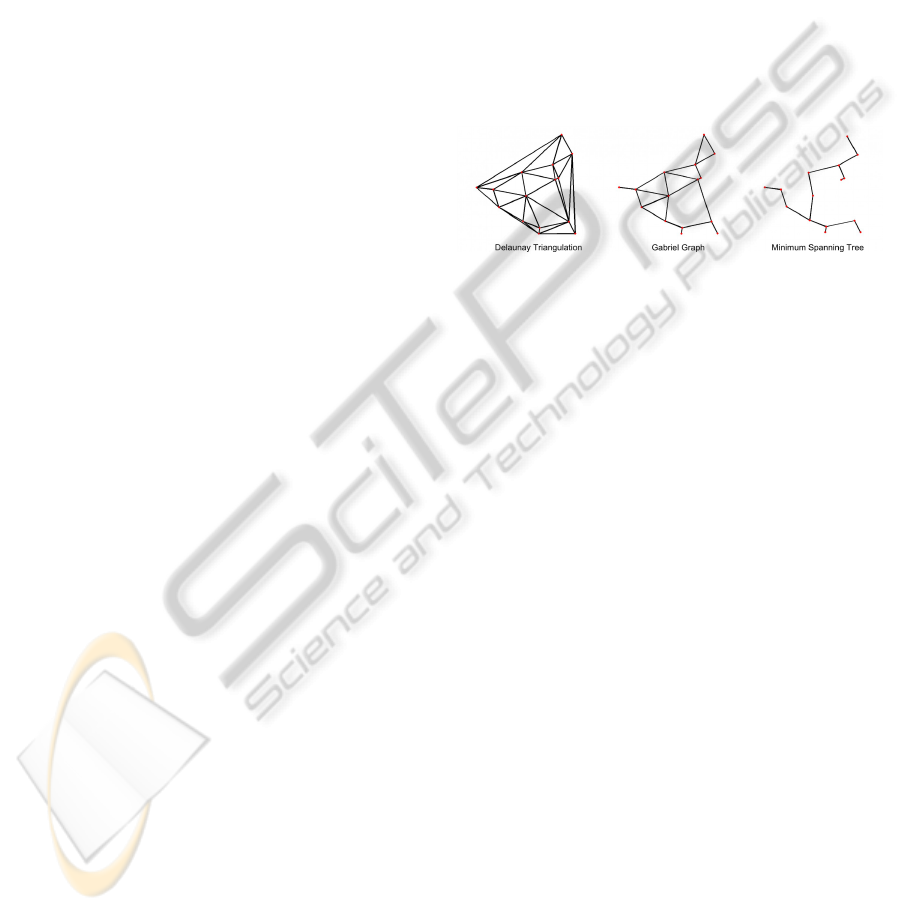
2.4 Graph Generation
After the segmentation of the cell nuclei, we have the
vertex set on which we build neighbourhood graphs.
Formally, a graph G = (V, E) is defined as a set V of
vertices of the graph and a set E of edges of the graph.
The methods developed uses the Voronoi diagram and
neighbourhood graphs stemming from the Voronoi
diagram: the Delaunay triangulation (DT), Gabriel
graph (GG) and the euclidean minimum spanning
tree (MST). Neighbourhood graphs are used in image
analysis to model geometric structures and connect-
edness. The edges define the neighbourhood relation
”is connected to” on the set of vertices. According
to Vincent (Vincent, 1989), and to Heijmans and Vin-
cent (Heijmans H., 1992), MST, GG and DT are very
interesting for studying proximity problems because
they are connected, unique (except for MST), they do
not depend on any parameter (e.g. a maximal distance
between objects or a minimal number of neighbours)
which implies that they are independent of scaling and
they are included into one another MST ⊆ GG ⊆ DT
[fig. 1], enabling a modelling of neighbourhood rela-
tionships of increasing strength.
Figure 1: Three neighbourhood graphs related to the
Voronoi diagram.
3 GLOBAL ANALYSIS - IMAGE
FIELD SCORE
In order to facilitate the subsequent high resolution
processing of the images we wish to detect regions
with low, medium and high density of cell nuclei. To
achieve this, a global analysis was performed to sepa-
rate the image field according to cell density.
• Low density regions: Some regions in the image
field are empty or contain very few nuclei.
• Medium density regions: These regions contain
many nuclei distributed quite evenly and belong-
ing mostly to free-lying cells. These are the re-
gions of biggest interest and they should be further
studied, the numerous nuclei allowing to build a
large database of features and measurements.
• High density regions: These regions contain a
very large number of nuclei or artifacts. Most of
the cells are overlapping and closely gathered in
clusters, some are folded and the dense regions
are more prone to be partly out of focus. The
nuclei can be deformed or barely visible. Their
segmentation can be very difficult, leading to use-
less measurements. Hence, special care should be
taken when studying these regions and thus it is
important to identify them.
ClusterDetectionandField-of-ViewQualityRating-AppliedtoAutomatedPap-smearAnalysis
357
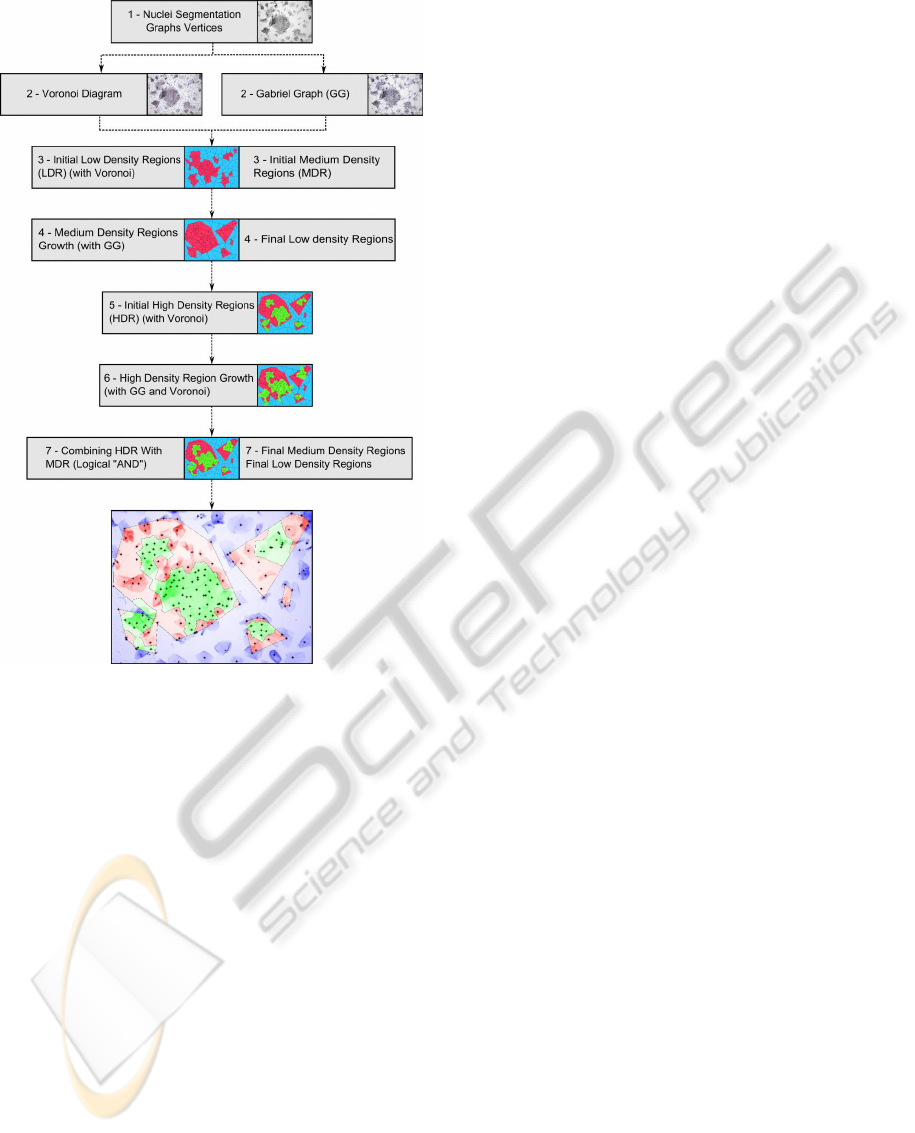
Figure 2: Global analysis method main steps.
The Global Analysis method uses both the Voronoi
Diagram and Gabriel Graph. The main steps of the
method are given in figure 2. Each step is further
discussed in the following sections. Throughout the
global analysis method, the parameters tuned (such as
thresholds) are normalized by the diameter of the im-
age, in order for the code to be adaptable to any image
size. All the parameters have been set by changing
values experimentally.
3.1 Determining the Low Density
Regions
Low density regions are defined as containing no or
few nuclei. Consequently, these regions hold very
few, spaced out, vertices, and are thus paved by large
Voronoi cells. In order to determine low density re-
gions, the cells in the Voronoi diagram were thresh-
olded according to their area. Each cell in the Voronoi
diagram with an area superior to a threshold is con-
sidered as belonging to a low density region. The rest
of the Voronoi cells are considered as belonging to
medium (or high) density regions [fig. 3 (a)].
3.2 Medium Density Regions Growth
After step 3 [fig. 2], we can observe on the Gabriel
graph that some vertices within the initial low density
regions are close (Euclidean distance) neighbours of
vertices situated inside the medium density regions.
These nuclei should thus be included in the medium
density regions, resulting in larger medium density re-
gion. This operation is repeated as long as vertices
are added. Once all the close-by vertices have been
added to the regions, we apply the convex hull on
each of the grown set of vertices, and thus obtain the
final medium density regions. The result is stored in a
binary image, where medium density regions are as-
signed the value 1 and low density regions (the rest of
the image) are assigned the value 0. The contours in
this binary image represent the external contours of
the final medium density regions. The internal con-
tours of the medium density regions will be the exter-
nal contours of the high density regions [fig. 3 (b)].
3.3 Initial High Density Regions
High density regions are defined as containing many
closely located nuclei. They are thus paved by small
Voronoi cells. In order to determine high density re-
gions, the cells in the Voronoi diagram were thresh-
olded according to their area. Each cell in the Voronoi
diagram with an area less than a threshold is consid-
ered as belonging to a high density region [fig. 3 (c)].
3.4 High Density Regions Growth and
Final Borders of
the High Density Regions
Likewise step 4 [fig. 2], after step 5, we can observe
on the Gabriel graph that some vertices outside the
initial high density regions are close (Euclidean dis-
tance) neighbours of vertices situated inside the high
density regions. These vertices are added to the high
density regions, but contrary to step 2, the operation is
just executed once, to avoid a too large growth of the
high density regions. Indeed, it was observed experi-
mentally that further iterations would result in adding
irrelevant vertices. Once these nearby vertices have
been added, one can notice that farther vertices be-
longing to high density regions are still considered
as belonging to medium density regions. To solve
this problem, we take into account the grey value of
the Voronoi cells situated in the neighbourhood of the
high density regions, from which we removed the nu-
clei that generated the Voronoi cell. If the mean grey
ICPRAM2013-InternationalConferenceonPatternRecognitionApplicationsandMethods
358

value is inferior to a threshold (dark Voronoi cell), or
if the standard deviation of the grey value is superior
to a threshold with a mean grey-value still quite low
(Voronoi cell with a dark part and a part of higher
intensity, often found at the border of a high density
region), the vertex that generated this Voronoi cell is
added to the high density regions vertices, resulting in
larger high density regions. This operation is repeated
as long as vertices are added. Indeed, high density re-
gions are often very dark due to the closeness of cells,
which justifies taking grey-value into account [fig. 3
(d)].
Once this step has been accomplished, we obtain
n sets of vertices belonging to the grown high density
regions. We extract the Voronoi cells generated by
these vertices, and store them in a binary image with
value 1. Value 0 is assigned to the rest of the image.
This binary image and the medium density regions bi-
nary image mentioned above are combined using the
logical operation AND [fig. 3 (e)]. The binary im-
age thus obtained is the final high density binary im-
age, where regions with value 1 represent high den-
sity regions. Indeed, high density regions should be
included in the regions that had previously been iden-
tified as medium dense, and not encroach upon low
density regions. The contours in this binary image
represent the contour of the final high density regions
[fig. 3 (f)].
3.5 Overview of the Results of
the Global Analysis
Figure 4 shows the results obtained with the global
analysis method, for a set of 30 images, 10 images
with mostly high density regions (HDR), 10 images
with mostly medium density regions (MDR) and 10
images with mostly low density regions (LDR).
Four ratios are calculated:
Ratio 1: Proportion of nuclei in MDR and nuclei in
LDR compared to nuclei in HDR.
nuclei in MDR + nuclei in LDR
nuclei in HDR + 1
(1)
Ratio 2: Proportion of MDR area compared to LDR
and HDR areas.
MDR Area
LDR Area + HDR Area
(2)
Ratio 3: Proportion of HDR area compared to LDR
and MDR areas.
Figure 3: Illustration of the Global Analysis main steps. (a)
Initial low density regions (LDR) (blue) and initial medium
density regions (MDR) (red). (b) Medium density regions
growth and final low density regions. (c) Initial high density
regions (HDR) (green). (d) High density regions growth.
(e) Combining HDR and initial MDR. (f) Final regions.
HDR Area
LDR Area + HDR Area
(3)
Ratio 4: Proportion of LDR area compared to HDR
and MDR areas.
LDR Area
HDR Area + MDR Area
(4)
The values obtained for the different ratios differ
according to the density of the nuclei in the image.
Ratio 1 increases when the nuclei density decreases.
For ratio 2 the proportion of MDR area compared to
LDR and HDR areas is much higher for medium den-
sity images than for low or high density images. Like-
wise, for ratio 3, the proportion of HDR area com-
pared to LDR and MDR areas is much higher for high
density images than for low or high density images,
and for ratio 4, the proportion of LDR area compared
to HDR and MDR areas is much higher for low den-
sity images than for medium or high density images.
ClusterDetectionandField-of-ViewQualityRating-AppliedtoAutomatedPap-smearAnalysis
359
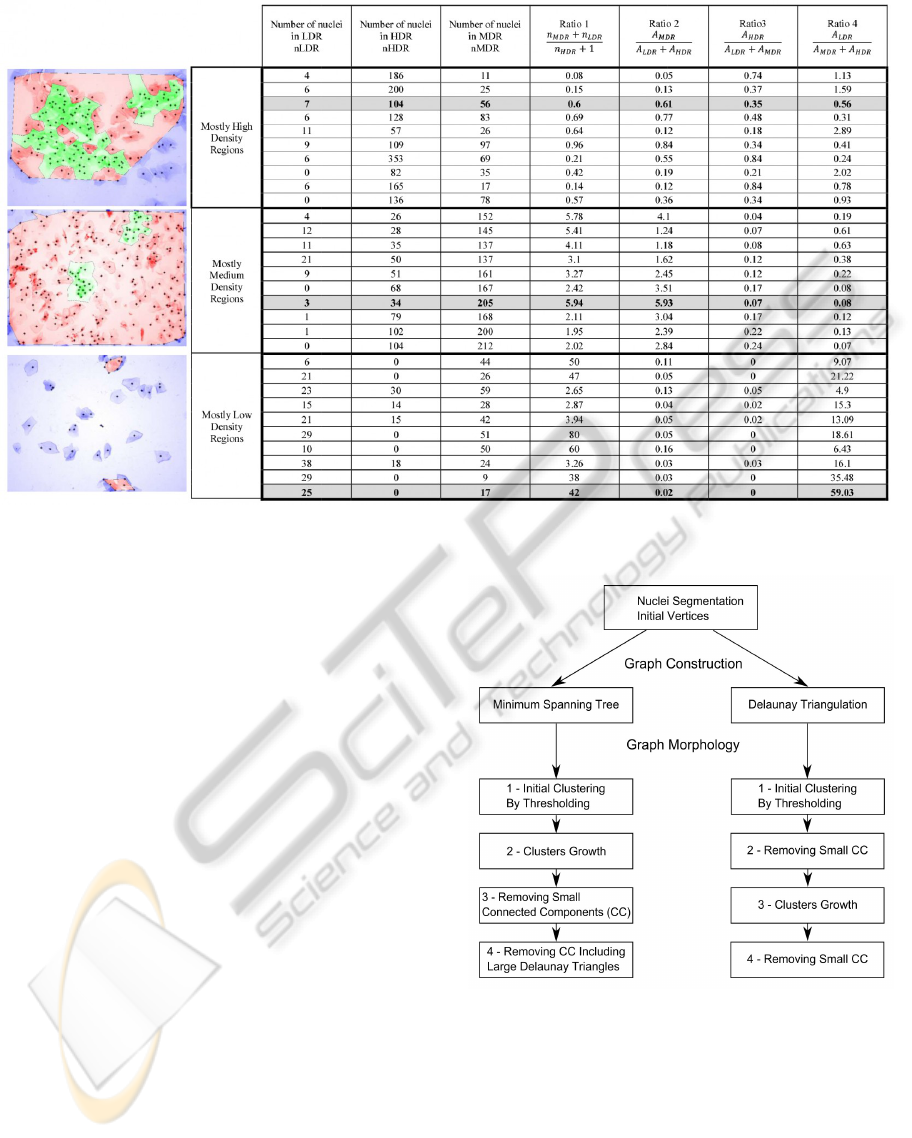
Figure 4: Overview of the results of the global analysis. The ratios values of the displayed images are highlighted.
4 DETECTING CELL CLUSTERS
USING GRAPHS
Cervical cell images usually contain single cells, clus-
ters of cells as well as artifacts. It is important to de-
fine the regions containing cells, clusters of cells, or
regions void of cells in order to reduce the area to be
studied. The detection of cell clusters in an image is
of interest for two reasons. First, cells have different
features when in a cluster, their nuclei can be over-
lapping or out of focus, resulting in the impossibility
to detect or to segment them properly. Special analy-
sis should be applied when studying cells in a cluster,
and therefore it is important to identify the position
of these clusters in the images. Secondly, the pres-
ence of cell clusters in the slide has a diagnostic value,
because malignant cells are more prone to stick to-
gether in clusters than normal cells. Thirdly, endome-
trial cells, which also should be detected, usually form
clusters. Graph theory is known for its ability to anal-
yse complex interactions and relationships in diverse
systems. Vertices correspond to the objects in the sys-
tem and the edges describe the neighbouring relations
between these objects. We have therefore used graph
theoretical methods to detect clusters. Throughout
the clustering method, the parameters tuned (such as
thresholds) are normalized by the diameter of the im-
age, in order for the code to be adaptable to any image
Figure 5: Proposed clustering methods main steps.
size. All the parameters have been set by changing
values experimentally.
4.1 Clustering the Euclidean Minimum
Spanning Tree
Two alternative approaches were tested to analyse the
clusters, which main steps are given in figure 5. The
first method was based on clustering the Euclidean
ICPRAM2013-InternationalConferenceonPatternRecognitionApplicationsandMethods
360

Minimum Spanning Tree (EMST).
4.1.1 Initial Clustering by Thresholding of the
Euclidean Minimum Spanning Tree
In order to obtain initial clusters, the edges in the
EMST are thresholded. Indeed, one feature character-
ising clusters in the EMST is their short edges. Each
edge in the EMST with a length superior to a thresh-
old t
Edges
is removed from the graph. The graph re-
sulting from the thresholding of the EMST is a forest
and the N
C
different connected components thus ob-
tained represent N
C
initial clusters.
4.1.2 Cluster Growth
From the previous step, we have obtained N
C
initial
clusters. The purpose of this step is to make each clus-
ter grow, by adding neighbouring edges similar (in
terms of length) to the edges already present in the ini-
tial cluster. A neighbouring edge is added if its length
divided by the average length of the edges in the initial
cluster is inferior to a threshold t
Edges−growth
. Once
every neighbouring edge has been visited, and pos-
sibly added, the resulting cluster is larger. Then the
operation is repeated on the resulting grown cluster,
as long as neighbouring vertices are added.
Once this step has been accomplished for each ini-
tial cluster that had been obtained after thresholding
of the EMST, we obtain N
C
clusters CG(grown clus-
ters). Edges and vertices have been added, and some
clusters CG are actually connected in the EMST. They
are then merged in order to identify each connected
components that will form our final set of clusters.
We end up with a number of clusters N
CG
≤ N
C
.
4.1.3 Removal of Small Connected Components
Connected components containing few vertices are
most of the time ”false clusters”, free-lying cells close
enough from each other to be mistaken as a cluster.
The clusters containing less than a threshold t
Comp
vertices are removed.
4.1.4 Removal of Connected Components
Including Large Delaunay Triangles
Clusters of cells are constituted of several cells very
close to each other (most of the time overlapping),
thus vertices (or nuclei) in clusters are also close to
each other and the Delaunay Triangles made up of
these vertices have a small area. As a result, con-
nected components that include Delaunay triangles of
large area are removed from the clusters list.
The result of the detection of clusters with the
EMST is shown for two cytology images in figure 6.
Figure 6: Detection of clusters using Euclidean Minimum
Spanning Tree. Black contours represent the clusters as de-
lineated for ground truth.
4.2 Clustering with the Delaunay
Triangulation
The second cluster analysis method was based on
clustering with the Delaunay triangulation (DT).
4.2.1 Initial Clustering by Thresholding of the
Delaunay Triangulation
In order to obtain initial clusters, the triangles in the
Delaunay triangulation were thresholded relatively to
their area and perimeter. Indeed, as noted previously,
vertices in clusters are close to each other, thus the
Delaunay triangles made up on these vertices have
small area and perimeter. Consequently, each trian-
gle in the Delaunay triangulation with an area supe-
rior to a threshold t
Area
and a perimeter superior to a
threshold t
Perimeter
is removed from the graph. The
new graph resulting from the thresholding of the De-
launay triangulation is a collection of connected com-
ponents. The N
C
different connected components thus
obtained represent N
C
initial clusters.
4.2.2 Removal of Small Connected Components
Before applying any transformation to the initial clus-
ters, we remove small connected components from
the initial clustering. Indeed, small connected com-
ponents are most of the time linking closely located
free-lying cells together and ”real clusters” are al-
ready represented by quite large connected compo-
nents after the initial clustering. Moreover, some of
the small connected components can grow consider-
ably, and then be kept as a cluster after step 4 (Re-
moval of small connected components after the clus-
ter growth), when it is in fact only a grouping of
closely located free-lying cells.
4.2.3 Delaunay Clusters Growth
N
C
initial clusters have been obtained from the pre-
vious step. The purpose of this step is to make each
cluster grow, by adding neighbouring triangles sim-
ilar (in terms of area and perimeter) to the triangles
ClusterDetectionandField-of-ViewQualityRating-AppliedtoAutomatedPap-smearAnalysis
361

already present in the initial cluster. A triangle is de-
fined as a neighbouring triangle of a cluster, if at least
one of its apexes is a vertex belonging to the cluster.
A neighbouring triangle is added to a cluster if its area
divided by the average area of the triangles in the ini-
tial cluster is inferior to a threshold t
Area−Growth
and if
its perimeter divided by the average perimeter of the
triangles in the initial cluster is inferior to a thresh-
old t
Perimeter−Growth
. Once every neighbouring trian-
gle has been visited, and possibly added, the resulting
cluster is larger. Then the operation is repeated on
the resulting grown cluster, as long as neighbouring
triangles are added. Once this step has been accom-
plished to each initial cluster, we obtain N
C
clusters
CG(grown clusters). Triangles, and so, vertices, have
been added, and some clusters CG are actually con-
nected in the Delaunay Triangulation. They are then
merged in order to identify each connected compo-
nents that will form our final set of clusters. We end
up with a number of clusters N
CG
≤ N
C
. As well as for
clustering with EMST, small connected components
are removed from the clusters list.
Results of the clustering with Delaunay triangula-
tion are shown in figure 7.
Figure 7: Detection of clusters using Delaunay Triangula-
tion.
4.3 Combining Clustering Methods
In this section we combine the results obtained by
clustering with the Euclidean minimum spanning tree
with the results obtained by clustering with the De-
launay triangulation.
The method employed returns a set of vertices
considered to be in clusters. Two ways of combin-
ing the methods have been considered. For the first
way, the vertices kept in this set are the vertices that
were considered as being in clusters both by the clus-
tering with EMST method and by the clustering with
DT method.
V
Combined−Intersect
= V
EMST
∩V
Delaunay
(5)
For the second way, the vertices kept in this set
are the vertices that were considered as being in clus-
ters by the clustering with EMST method or by the
clustering with DT method. Figure 8 illustrates the
Figure 8: Detection of clusters using the intersection of
clustering methods results.
Figure 9: Detection of clusters using the union of clustering
methods results.
results obtained while detecting clusters using the in-
tersection of above-presented methods, and figure 9,
using the union.
V
Combined−U nion
= V
EMST
∪V
Delaunay
(6)
From both sets of vertices VCombined, we iden-
tify the different connected components (connected in
the Delaunay triangulation), in order to separate the
different clusters. We obtain several sets of vertices,
each set containing the vertices of a specific cluster.
4.4 Results
4.4.1 Ground Truth
The performance of the clustering methods was eval-
uated relative to manual definition of the clusters. A
Graphical User Interface (GUI) was developed to per-
mit a user to delineate regions containing clusters in
a set of images. A cervical cell image analysis expert
used the GUI to trace the clusters boundaries. A to-
tal of 48 images, belonging to different samples, have
been labelled and used to evaluate and compare the
methods. The accuracy of the methods is calculated
using the 48 images, containing a total of 2307 nu-
clei labelled as belonging to a cluster and 8116 nuclei
labelled as not belonging to a cluster.
4.4.2 The Cellgraph Method
In order to compare our method to the cellgraph based
method developed by Chandran et al. (Chandran
et al., 2012), we implemented that method using the
centroids of the segmented nuclei as a set V of ver-
tices of the graph. Some results using the cellgraph
ICPRAM2013-InternationalConferenceonPatternRecognitionApplicationsandMethods
362

method are illustrated in figure 10. The cellgraph
method, as developed by Chandran et al., uses the
Waxman Model (Waxman, 1988), in which edges are
probabilistically assigned between vertices and the
probability of linking a pair of vertices v and w with
an edge decreases with the increase in the Euclidean
distance between them.
Figure 10: Detection of clusters using the cellgraph method.
4.4.3 Evaluation
Each method classifies nuclei (or vertices) as belong-
ing to a cluster or not belonging to a cluster. To mea-
sure the performance of each method, we calculate
the sensitivity and specificity defined as follow:
Speci f icity =
True Negative
True Negative + False Positive
× 100
(7)
Sensitivity =
True Positive
True Positive +False Negative
× 100
(8)
where a positive refers to a nucleus belonging to a
cluster and a negative, to a nucleus belonging to a
free-lying cell. True or false states the correctness of
the classification in one of the two above mentioned
groups.
The specificity represents the probability that the
nucleus is classified as not in a cluster given that the
nucleus is indeed not in a cluster.
The sensitivity represents the probability that the
nucleus is classified as in a cluster given that the nu-
cleus is indeed in a cluster.
Figure 11 shows the resulting specificity and sen-
sibility for each method. As expected, specificity in-
creases and sensitivity decreases when using the in-
tersection of both methods and conversely, specificity
decreases and sensitivity increases when using the
union of both methods. Segmentation issues (nuclei
that had not been detected or that had been removed
after classification, or artifacts that had been mistaken
as nuclei) often causes the methods to be imprecise
and improving the segmentation would benefit to the
clustering results. It is also difficult to define precisely
a cluster, and it was observed that some clusters de-
tected by the method, which had not been marked as
ground truth clusters, could actually be clusters.
Figure 11: Evaluation of the clustering methods with 95%
confidence intervals.
5 CONCLUSIONS
In this paper, we presented a low resolution cell find-
ing system for fields-of-view quality rating and clus-
ter detection in bright-field microscope images of cer-
vical cells. Neighbourhood graphs have been fit to
the image nuclei in order to model neighbourhood re-
lationships between cells. Transformations on such
graphs resulted in the detection of cell clusters and
the delineation of regions with varying degree of cell
density. The evaluation of our clustering methods in
term of sensitivity and specificity, as well as a com-
parison to a related work in the literature shows that
our approach is accurate, effective and relevant for the
detection of cells in clusters. We believe that the per-
formance of our methods can be further increased by
improving nuclei segmentation results.
REFERENCES
Bengtsson, E. (2005). Computerized cell image processing
in healthcare. In Choi, HK, editor, Healthcom 2005:
7th International Workshop on Enterprise Networking
and Computing in Healthcare Industry, Proceedings,
pages 11–17, 345 E 47TH ST, New York, NY 10017
USA. Korea Multimedia Soc; IEEE; Inje Univ; Busan
Convent Bur, IEEE. 7th International Workshop on
Enterprise Networking and Computing in Healthcare
Industry (HEALTHCOM 2005), Busan, South Korea,
JUN 23-25, 2005.
Chandran, S. P., Byju, N. B., Deepak, R. U., Kumar, R. R.,
Sudhamony, S., Malm, P., and Bengtsson, E. (2012).
Cluster detection in cytology images using the cell-
graph method. In IT in Medicine and Education
(ITME), 2012 International Symposium on.
Elsheikh, T. M., Austin, R. M., Chhieng, D. F., Miller, F. S.,
Moriarty, A. T., and Renshaw, A. A. (2012). American
society of cytopathology workload recommendations
for automated pap test screening: Developed by the
productivity and quality assurance in the era of auto-
mated screening task force. Diagnostic Cytopathol-
ogy, 1:1.
ClusterDetectionandField-of-ViewQualityRating-AppliedtoAutomatedPap-smearAnalysis
363

Grohs, H. and Husain, O., editors (1994). Automated Cervi-
cal Cancer Screening. IGAKU-SHOIN Medical Pub-
lishers, Inc.
Heijmans H., V. L. (1992). Mathematical Morphology in
Image Processing. CRC Press; First Edition edition,
New York.
Moshavegh, R., Ehteshami, B., Mehnert, A., Sujathan, K.,
Malm, P., and Bengtsson, E. (2012). Automated seg-
mentation of free-lying cell nuclei in pap smears for
malignancy-associated change analysis. In Engineer-
ing in Medicine and Biology Society (EMBC), 2012
Annual International Conference of the IEEE.
Nordin, B. (1989). The development of an automatic pre-
screener for the early detection of cervical cancer: Al-
gorithms and implementation. PhD thesis, Uppsala
University.
Papanicolaou, G. N., Traut, H. F., and Stanton A. Friedberg,
M. (1943). Diagnosis of uterine cancer by the vaginal
smear. Oxford University Press, New York.
Poulsen, R. S. (1973). Automated prescreening of cervical
cytology specimens. PhD thesis, Department of Elec-
trical Engineering, McGill University.
Raymond, E., Raphael, M., Grimaud, M., Vincent, L., Bi-
net, J. L., and Meyer, F. (1993). Germinal center anal-
ysis with the tools of mathematical morphology on
graphs. Cytometry, 14(8):848–61.
Saslow, D., Solomon, D., Lawson, H. W., Killackey, M.,
Kulasingam, S. L., Cain, J., Garcia, F. A. R., Moriarty,
A. T., Waxman, A. G., Wilbur, D. C., Wentzensen, N.,
Downs, L. S., Spitzer, M., Moscicki, A.-B., Franco,
E. L., Stoler, M. H., Schiffman, M., Castle, P. E., and
Myers, E. R. (2012). American cancer society, amer-
ican society for colposcopy and cervical pathology,
and american society for clinical pathology screening
guidelines for the prevention and early detection of
cervical cancer. American Journal of Clinical Pathol-
ogy, 137(4):516–542.
Vincent, L. (1989). Graphs and mathematical mor-
phology. Signal Processing, 16(4):365 – 388.
¡ce:title¿Special Issue on Advances in Mathematical
Morphology¡/ce:title¿.
Waxman, B. (1988). Routing of multipoint connections.
Selected Areas in Communications, IEEE Journal on,
6(9):1617 –1622.
WHO/ICO Information Centre on HPV and Cervical Can-
cer (HPV Information Centre) (2012). Human papillo-
mavirus and related cancers in world. summary report
2010.
Zuiderveld, K. (1994). Graphics gems iv. chapter Contrast
limited adaptive histogram equalization, pages 474–
485. Academic Press Professional, Inc., San Diego,
CA, USA.
ICPRAM2013-InternationalConferenceonPatternRecognitionApplicationsandMethods
364
