
Semantic Interoperability Solution for Multicentric Breast Cancer
Trials at the Integrate EU Project
Sergio Paraiso-Medina
1
, David Perez-Rey
1
, Raul Alonso-Calvo
1
, Brecht Claerhout
2
,
Kristof de Schepper
2
, Philippe Hennebert
3
, Jérôme Lhaut
3
, Jasper Van Leeuwen
4
and Anca Bucur
4
1
Grupo de Informática Biomédica, DIA & DLSIIS, Facultad de Informática,
Universidad Politécnica de Madrid, Campus de Montegancedo S/N, 28660 Boadilla del Monte, Madrid, Spain
2
Custodix NV, Kortrijksesteenweg 214b3, 9830 Saint-Martens-Latem, Belgium
3
Institut Jules Bordet, 121 boulevard de Waterloo, 1000 Brussels, Belgium
4
Phillips Research, Healthcare Information Management, High Tech Campus 34, 5656 AE Eindhoven, The Netherlands
Keywords: Semantic Interoperability, Clinical Trials, SNOMED, HL7, INTEGRATE Project, Breast Cancer.
Abstract: The introduction of –omic information within current clinical treatment is one of the main challenges to
transfer the huge amount of genomic-based results. The number of potential translational clinical trials is
therefore experiencing a dramatic increase, with the corresponding increment on patient variability. Such
scenario requires a larger population to recruit a minimum set of patients that may involve multi-centric
trials, with associated challenges on heterogeneous data integration. To ensure sustainability on clinical trial
management, semantic interoperability is one of the main goals addressed by international initiatives such as
the EU funded INTEGRATE project: “Driving Excellence in Integrative Cancer Research”. This paper
describes the approach adopted within an international research initiative, providing a homogeneous
platform to manage clinical information from patients on breast cancer clinical trials. Following the project
“leitmotif” of reusing standards supported by a large community, we have developed a solution providing a
common data model (i.e. HL7 RIM-based), a biomedical domain vocabulary (i.e. SNOMED) as core dataset
and resources from the semantic web community adapted for the biomedical domain. After one year and a
half of collaboration, the INTEGRATE consortium has been able to develop a solution providing the
reasoning capabilities required to solve clinical trial patient recruitment. The next challenge will be to
extend the current solution to support a cohort selection tool allowing prospective analysis and predictive
modeling.
1 INTRODUCTION
Current oncology treatments are introducing a large
number of new variables to current clinical
guidelines. Molecular tests, in addition to traditional
clinical variables, are inducing an explosion in the
number of potential –if not always actual– clinical
trials. In addition, the high specificity of eligibility
criteria, especially molecular criteria, focusing on
sometimes rare gene mutations, makes patient
recruitment more difficult, increasing the need for
multi-centric and international initiatives.
Information systems with complex data from
different institutions have to deal with additional
heterogeneities on different biomedical vocabularies,
data models, security procedures, legislation, etc.
Integration processes, carried out manually until
now, are becoming less and less manageable with
the dramatic increase of variables and centers
involved.
Within such scenario, interoperability among
different systems (i.e. communication and
understanding of data transferred) is essential to
provide a sustainable solution. This work has been
carried out within the three-year EU funded
INTEGRATE (INTEGRATE, 2012) research
project. The main goal of the INTEGRATE platform
is to provide solutions to clinical researchers and the
pharmaceutical industry for sharing of data and
knowledge, support for molecular testing scenario
for patient enrolment in trials, querying trial data,
and building and sharing of predictive models for
response to therapies.
34
Paraiso-Medina S., Perez-Rey D., Alonso-Calvo R., Claerhout B., de Schepper K., Hennebert P., Lhaut J., Van Leeuwen J. and Bucur A..
Semantic Interoperability Solution for Multicentric Breast Cancer Trials at the Integrate EU Project.
DOI: 10.5220/0004223400340041
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2013), pages 34-41
ISBN: 978-989-8565-37-2
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

This paper describes the approach proposed to
provide semantic interoperability to the
INTEGRATE platform. The Background section
describes previous projects facing similar
challenges. Then, the Semantic Interoperability
Layer section presents the required components and
the query expansion mechanism implemented to
retrieve data with semantic reasoning capabilities.
The Results section describes preliminary
performance tests using real clinical data for patient
recruitment. And finally, the Conclusions section
describes the main contribution of this work and
future lines.
2 BACKGROUND
The latest advances on breast cancer research have
produced a wealth of data. To obtain a proportional
increment of knowledge, previous projects have
been focused on cancer research studies and
heterogeneous biomedical integration such as
caBIG, i2b2, OMOP and ACGT.
The cancer Biomedical Informatics Grid (caBIG)
(Eschenbach von and Buetow, 2006) is an open
source information network deployed in 2003,
allowing cancer researchers to share tools, data and
applications, and to agree upon common standards
and needs. It is based on Open Grid Services
Architecture (OGSA and OGSA-Data Access
Integration, OGSA-DAI (Antonioletti et al., 2005))
and the open source grid computing project, Globus
Toolkit from Globus Alliance (Globus, 2012).
Applications developed for caBIG are highly
dependent on the GRID-based middleware, which
difficult reusing applications outside the caBIG
framework.
Informatics for Integrating Biology and the
Bedside (i2b2) is a framework based on the
Research Patient Data Registry developed at the
Massachusetts General Hospital (Murphy Shawn N
et al., 2010). i2b2’s main goal is to allow researchers
to use the clinical data for discovery research. It is
designed as a set of services, denominated cells,
which fit together in an integrated environment
(called a hive). Every cell is a SOA service like a
file repository, ontology management, data
repository, etc. The data repository cell is designed
as the data warehouse to provide the information of
the users. i2b2 includes an ontology cell to define
the vocabulary, but this vocabulary only allows one
type of relationship, thus medical ontologies like
SNOMED CT (SNOMED, 2012) cannot fully stored
within this implementation.
Observational Medical Outcomes Partnership
(OMOP) was a clinical project to analyze healthcare
databases for studying issues and effects of medical
products (Stang et al., 2010). One of the advantages
of the OMOP is the simple data model. But similar
to i2b2, it has some problems including an ontology
vocabulary like SNOMED CT in the model. In fact,
OMOP only provides a dictionary that performs the
mapping between different data sources and the
database.
Projects described above have been very
valuable in obtaining practical results, but few of
them have exploited the benefits of the current
semantic web tools. In this context, emerged ACGT
(FP6-2005-IST-026996), an EU funded Project
devoted to the development of a technological
platform for supporting clinical trials on cancer
(Martin et al., 2011). The platform included an ad
hoc ontology built specifically for ACGT, i.e. the
Master Ontology on Cancer (MO) (Brochhausen et
al., 2011), as data model and domain vocabulary. A
semantic mediation layer was developed to
dynamically translate queries in terms of the MO to
the concrete schemas of data sources. While
providing an efficient layer for the integrated access
to a set of disparate resources, the complexity of the
MO (over a thousand classes with hundreds of
properties), hindered its use as schema for users to
build meaningful queries.
Further resources, developed within the semantic
web community, have been adapted to the
biomedical domain. Nowadays, there are classifiers
to solve the management of large ontologies such as
SNOMED CT, CEL (Baader et al., 2006),
SNOROCKET (Lawley and Bousquet, 2010) and
ELK (Kazakov et al., 2012) instead of general
purpose reasoners such as Pellet (Parsia and Sirin,
2004), Fact++ (Tsarkov and Horrocks, 2003) or
Hermit (Shearer R et al., 2008). Classifiers are
reasoners with specific algorithms of inferring
optimal for certain types of ontologies. Comparing
classifiers to deal with SNOMED CT, ELK was
more efficient than CEL and SNOROCKET
(Kazakov et al., 2011). And although classified
ontologies are around 75% lighter than the original,
semantic repositories are required to efficiently store
them. Examples include Sesame (Broekstra et al.,
2002), Virtuoso (Erling and Mikhailov, 2009) or
OWLIM (Kiryakov et al., 2005).
These repositories are used in the semantic web
to store data represented in markup languages, with
efficient search engines to extract domain
knowledge in terms of relationships.
SemanticInteroperabilitySolutionforMulticentricBreastCancerTrialsattheIntegrateEUProject
35
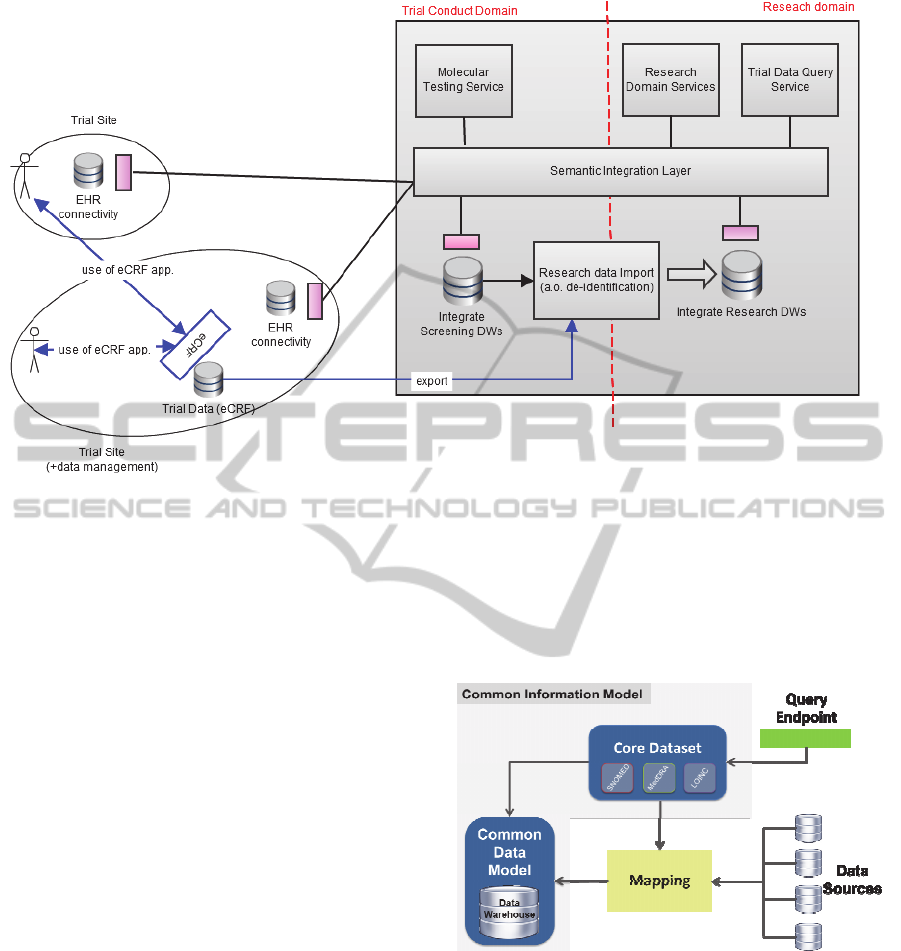
Figure 1: Overview of the INTEGRATE platform general architecture. eCRF: Electronic Case Report Form; EHR:
Electronic Health Record; DWs Datawarehouses.
The review of the state of the art suggested that
semantic resources can still be largely exploited to
improve the semantic interoperability of integrative
solutions.
3 SEMANTIC
INTEROPERABILITY LAYER
Semantic interoperability among applications and
tools is an essential requirement to achieve the main
goal of the INTEGRATE project, i.e. data sharing
for breast cancer clinical trials. The general
architecture of the proposed platform includes
different services, presented in Figure 1.
The INTEGRATE platform has to deal with two
main scenarios, each belonging to one of two
operational domains: (i) Trial Conduct Domain and
(ii) Research Domain. In the Trial Conduct Scenario,
patients are recruited into clinical trials from each
site, and relevant information from CRFs and EHRs
sources is homogeneously represented. In this
scenario, patients should remain identified until the
end of the clinical trial, and therefore, information
should remain distributed at each site. The Research
Domain, however, deals with encoded data to allow
researchers to perform retrospective analysis of
multiple clinical trials or predictive modeling.
To provide homogeneous access to different data
sources, the semantic interoperability layer should
provide a Common Information Model (CIM) to
represent the information. Thus, a common query
endpoint can be provided to retrieve semantically
uniform data. Components required for this task are
shown in Figure 2.
Figure 2: Semantic interoperability layer components.
The CIM proposed for the INTEGRATE
platform semantic layer is comprised by two
components: (i) the Core Dataset (CD) and (ii) the
Common Data Model (CDM). CDM refers to the
schema of the Data Warehouse and CD is the
domain vocabulary of the INTEGRATE platform.
This vocabulary, previously transformed into a
XML-based ontology representation language, is
stored in a semantic web repository. The CD will be
used to extract domain knowledge to retrieve data
HEALTHINF2013-InternationalConferenceonHealthInformatics
36
stored within the CDM.
In the following sections, a detailed description
of components previously mentioned is provided.
The query expansion subsection describes the
proposed method to semantically retrieve
information from the CDM by considering relations
contained within the domain vocabulary (CD).
3.1 Common Data Model
CDM proposed at INTEGRATE is the structure of
the Data Warehouse, and a wrapper that offers a
SPARQL endpoint. The CDM resolves
heterogeneity problems in different data sources.
Therefore, the CDM acts as the central data model
of the semantic interoperability layer. Information
from the different data sources is stored in different
Data Warehouses (distributed across the
institutions). This information is extracted,
transformed and loaded into those Data Warehouses
by the mapping tools.
HL7 RIM was the standard selected to develop
the common data model for the INTEGRATE
platform. HL7 RIM (HL7, 2012) includes most
common healthcare domains and serves as a general
data model for healthcare administrative and clinical
information. A relational database, based on HL7
RIM was therefore developed. Messages, documents
and rules conforming to that model are also defined
by HL7.
SPARQL was the query language selected to
query the information loaded in the Data
Warehouse. It is “de facto” standard, and W3C
recommendation, for querying RDF in the semantic
web. SPARQL also facilitates the federation of
queries in different data sources. To obtain a
SPARQL endpoint the D2R Server (Bizer C and
Cyganiak R, 2006) has been applied to publish
relational databases on the Semantic Web.
3.2 Core Dataset
Since the HL7 RIM data model does not specify the
vocabulary to be used for semantic representation of
concepts, a choice of domain ontology had to be
made to act as a “lingua franca” and to facilitate
extracting and exchanging information. We
considered different candidates such as SNOMED
CT, LOINC (McDonald CJ et al., 2003) or
MedDRA (Brown EG et al., 1999). SNOMED CT
(one of the largest medical ontology, developed,
distributed and maintained by IHTSDO) was
selected for the INTEGRATE platform. SNOMED
CT also provides mechanisms for identifying post-
coordinated concepts and adding new concepts with
extensions.
SNOMED CT consists of over 400.000 medical
concepts, with about one million descriptions and
more than one million relationships. Therefore, this
large amount of information implies a great
complexity to be managed. We used classifiers to
infer implicitly stated knowledge from explicitly
represented information, thereby eliminating
inconsistencies, incongruities and all types of
information not expected.
Among available classifiers, ELK has been
selected to classify the CD, filtering required
relationships to improve performance. SNOMED
CT was firstly transformed into the Ontology Web
Language (OWL) (McGuinness DL and Van
Harmelen F, 2012). Once a classified version of
SNOMED CT was obtained, it was necessary to use
a semantic repository to store it. Sesame was
selected in this case.
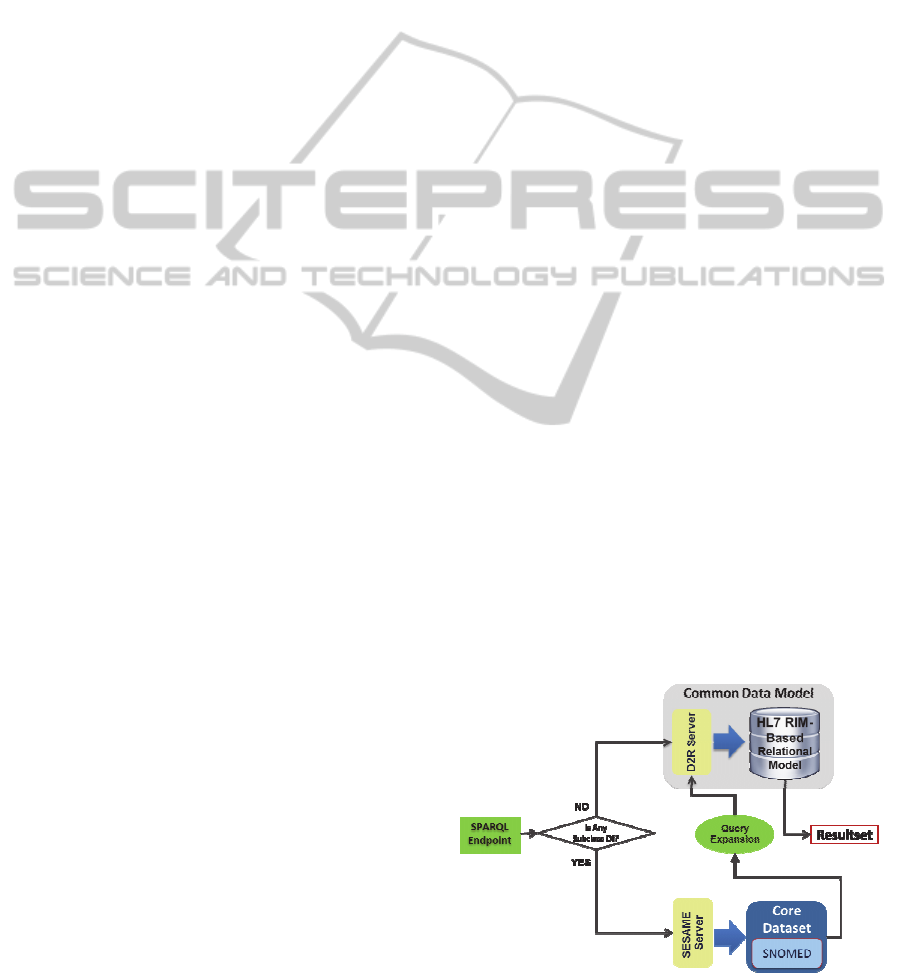
3.3 Query Expansion
To retrieve semantically uniform information from
the CDM, a query expansion method has been
proposed. The objective in this case is to exploit
relationship information contained within the CD
when querying the platform. A data flow of this
process is showed in Figure 3.
The semantic interoperability layer receives a
SPARQL query. If the original query does not need
query expansion for any concept, then the query is
sent directly to the CDM. If the query requires to be
expanded, it is enriched with concepts from the CD
and sent to the CDM. The CD Sesame repository
receives the concept that may require expansion and
includes the corresponding information in the
original query. Queries are enriched from “is_a” tree
structures as showed in Figure 5.
Figure 3: INTEGRATE query expansion method.
SemanticInteroperabilitySolutionforMulticentricBreastCancerTrialsattheIntegrateEUProject
37
Sesame returns the subclass information of an
Entity in terms of “is_a” relationships. Then, the
Conjunctive Normal Form (CNF) from this concept
is used to expand the original query as shown below.
CONCEPT = Entity OR Subclass
11
OR
Subclass
12
OR … OR Subclass
nm
In post-coordination cases, the original query
considers concepts that have to be expanded and the
relationship among them (represented by CDM
structure). Similar to simple query expansion, the
CD Sesame repository receives the concepts that
may require expansion and includes the
corresponding information in the original query
from a tree structure such as Figure 8.
A CNF of the post-coordination concept is then
built (as shown below) and sent to the CDM as
previously explained. Thus,
(CONCEPT = Entity A OR SubclassA
11
OR
SubclassA
12
OR … OR SubclassA
nm
)
AND
(RELATIONSHIP = Entity B OR
SubclassB
11
OR SubclassB
12
OR … OR
SubclassB
lk
)
Finally, the expanded query is sent to the D2R
wrapper of the CDM and executed in the data
warehouse.
4 RESULTS
A prototype of the semantic interoperability layer,
implementing the query expansion method was
developed using SOAP services and JAVA. Data
constructed based on actual data from 50 patients
from TOP (TOP, 2012) and NeoALTTO (Neo-
ALTTO, 2012) clinical trials, were loaded into the
HL7 RIM-based common data model and used to
test the INTEGRATE semantic interoperability
layer. Queries were built to match eligibility criteria
(EC) from those clinical trials.
The following eligibility criteria did not require
query expansion:
Inclusion criterion 2: Age of patient <= 70 years
Inclusion criterion 3: Female patient
Inclusion criterion 6: Patients with fixed samples
from the primary tumor
Inclusion criterion 8: Patients who signed the
informed consent
Inclusion criterion 10a: Patients with
ANC>=1500 mm
3
Inclusion criterion 10b: Patients with GOT <=
1.5N
Inclusion criterion 10c: Patients with
GPT>=1.5N
These EC queries were built using SPARQL and
directly executed on a D2R server that was mapped
onto the INTEGRATE CDM.
The following eligibility criteria did require
simple query expansion or post-coordination query
expansion:
Exclusion criterion 6: Patients with previous
treatment with anthracyclines
Exclusion criterion 1: Patients with metastatic
breast cancer
Inclusion criterion 1: Patients with
Histologically-confirmed breast cancer
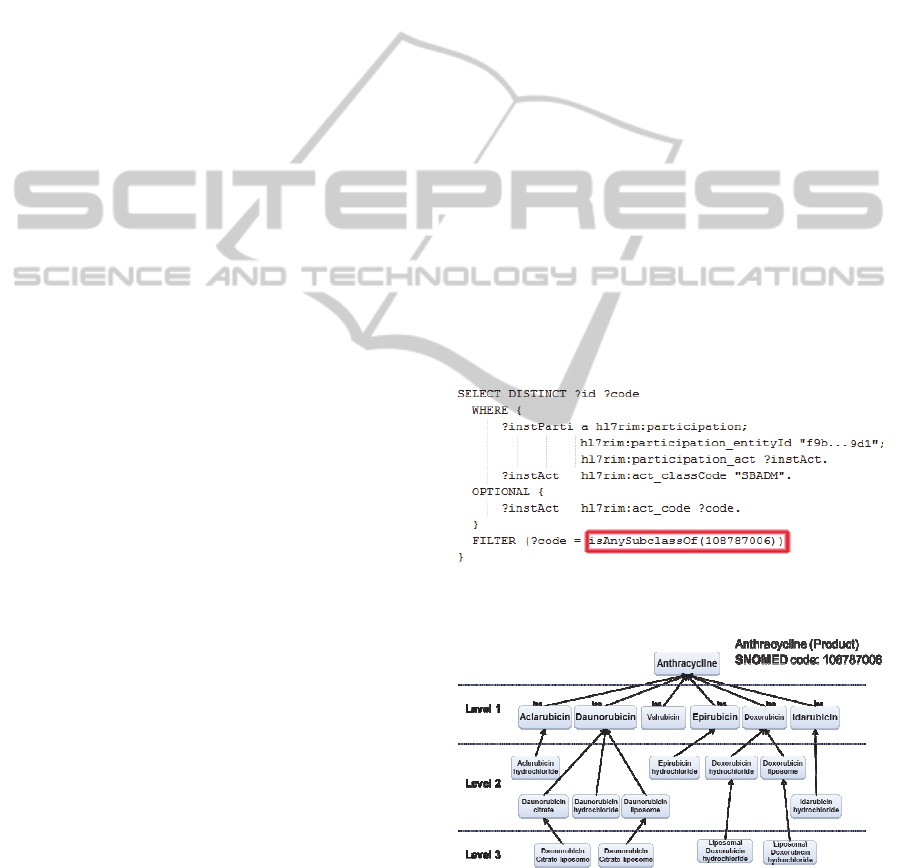
The first of these eligibility criteria required to
retrieve information about patient with previous
treatment with Anthracyclines. But it is unlikely that
actual data contains any drug labeled as
Anthracyclines, since it is a family of drugs. In our
case, data contained subclasses of Anthracyclines
such as Daunorubicin, Epirubicin, Idarubicin, etc.
Instead, the query given to the semantic
interoperability layer is showed in Figure 4.
Figure 4: Original SPARQL query on Anthracycline
eligibility criterion.
Figure 5: Anthracycline tree structure.
This SPARQL query, with a function called
isAnySubclassOf, is sent to the semantic
interoperability layer. This function triggers the
query expansion method with the required concept.
HEALTHINF2013-InternationalConferenceonHealthInformatics
38
Then, the CD retrieves the subclasses of
Anthracycline treatment in SNOMED, obtaining the
following tree structure (Figure 5).
A CNF is then built with the previous tree
structure. This CNF replaces the function
isAnySubclassOf in the given query, which is then
sent to the CDM, as shown in Figure 6.
Figure 6: Final (expanded) SPARQL query on
Anthracyclines eligibility criterion.
Thereby, information about patients with any
kind of anthracyclines treatment is retrieved from
the CDM.
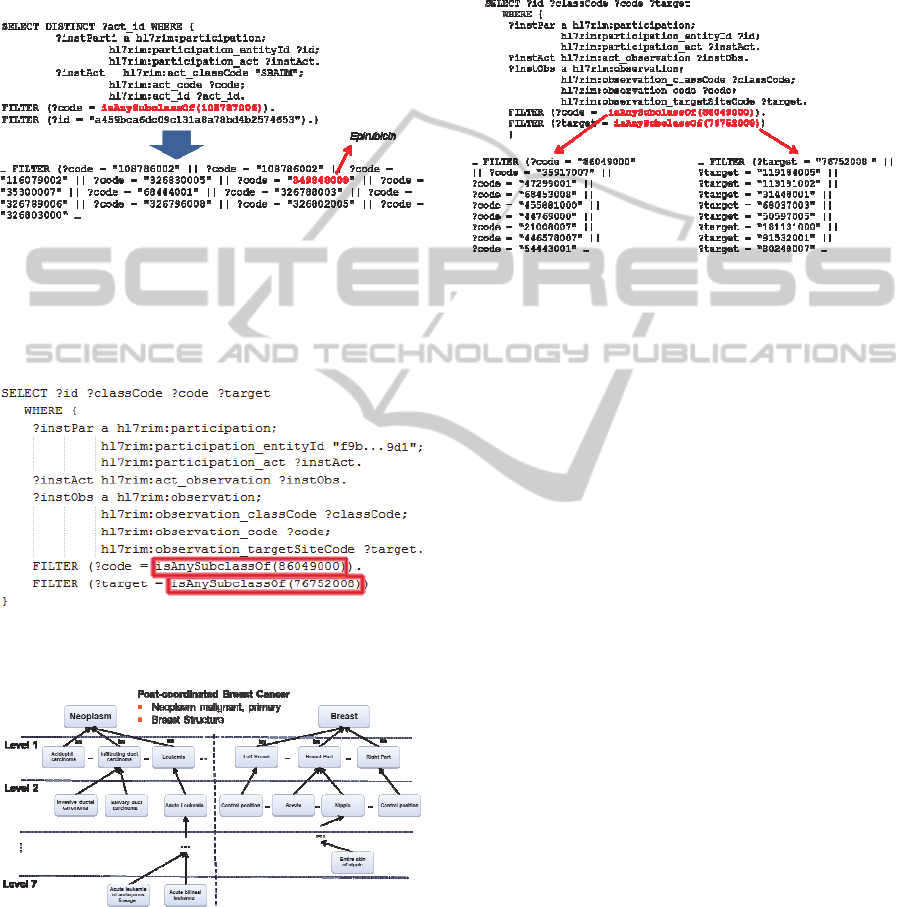
Figure 7: Original SPARQL query on breast cancer (post-
coordinated) eligibility criterion.
Figure 8: Breast Cancer tree structures.
Exclusion criterion number 1 and inclusion
criterion number 1 consult information relates to the
post-coordinated concept breast cancer. This term
links the SNOMED CT concept of Neoplasm with
its target site, in this case Breast. Figure 7 shows the
SPARQL query provided to the semantic
interoperability layer, where the function
isAnySubclassOf triggers the query expansion
method with the SNOMED CT code for Neoplasm
and the SNOMED CT code for the target site
Breast.
Figure 9: Final (expanded) SPARQL query on Breast
Cancer (post-coordinated) eligibility.
As it can be seen in Figure 8 and Figure 9, the
Neoplasm concept and its target site Breast are
expanded by two tree structures. Both structures
replaced the concepts of Breast Cancer in the
original query, by any subclass of Neoplasm (x
subclasses) and Breast (and subclasses). The
expanded query was sent to the CDM, retrieving
results of patients, and fulfilling the initial semantic
capabilities required by a patient recruitment tool for
clinical trials.
5 CONCLUSIONS
This paper has presented the semantic
interoperability approach developed within the
INTEGRATE project. The main challenge was to
provide a homogenous and powerful solution to
facilitate the collaboration among a complex set of
tools required to manage post-genomic clinical trials
on breast cancer.
After a comprehensive review of the literature in
the area, we identified two main issues that have not
been always addressed in previous efforts: (i) solve
post-genomic clinical trial heterogeneities by (ii)
exploiting semantic web technologies. Semantic web
technologies have been extensively developed
during the last years, together with data models and
domain ontologies that required long periods of time
to develop. Specific characteristics of the biomedical
research nowadays require advanced methods to
solve basic problems of interoperability. These
interoperability issues are essential to enhance
SemanticInteroperabilitySolutionforMulticentricBreastCancerTrialsattheIntegrateEUProject
39

patient care, by supporting integrative prospective
analysis and predictive modeling over multi-centric
datasets.
Patient data from multi-centric and international
clinical trials have been used to test the proposed
solution, suggesting the suitability of the proposed
solution. Next steps of the project will be focused
on: (i) testing performance for large amounts of
patient data (cohort selection), (ii) a cache
implementation to support large reasoning, (iii)
formalization of post-coordination related reasoning
and (iv) extension of such reasoning with new types
of relationships for the query expansion method.
After a year and a half of the INTEGRATE joint
effort, we have already undertaken essential issues
to improve the management of post-genomic clinical
trials. We have adapted and successfully applied
semantic web technologies to the complex domain
of biomedical research. The next steps aim to
support a crucial challenge nowadays, enhancing the
translation of –omic research to improve clinical
practice in oncology patients.
ACKNOWLEDGEMENTS
The present work has been funded by the European
Commission through the INTEGRATE project
(FP7-ICT-2009-6-270253).
REFERENCES
Antonioletti, M., Atkinson Malcolm, Baxter R., Borley A.,
Chue N. P., Collins B., Hardman N., et al., 2005. The
Design and Implementation of Grid Database Services
in OGSA-DAI. In Concurrency and Computation:
Practice and Experience, Volume 17, Issue 2-4, Pages
357-376.
Baader F., Lutz C and Suntisrivaraporn B., 2006. CEL – a
polynomial.time reasoner for life science ontologies.
In Lectures Notes in Computer Science, Volume
4130/2006, 287-291, DOI: 10.1007/11814771_25.
Bizer C. and Cyganiak R., 2006. D2R Server – Publishing
relational databases on the semantic web. In the 5
th
International Sematic Web Conference (ISWC).
Brochhausen M., Spear A. D., Cocos C., Weiler G.,
Martin L., Anguita A., Stenzhorn H., Daskalaki E.,
Schera F., Schwarz U., Sfakianakis S., Kiefer S., Dörr
M., Graf N. and Tsinakis M. ,2011. The ACGT Master
Ontology and its applications--towards an ontology-
driven cancer research and management system. In J
Biomed Inform; 44(1): 8-25.
Broekstra J, Kampman A and van Harmelen F. 2002.
Sesame: An Architecture for Storing and Querying
RDF and RDF Schema. In: Proceedings of the First
International Semantic Web Conference (ISWC2002).
Number 2342 in Lecture Notes in Computer Science
(LNCS), Springer-Verlag (2002) 54–68.
Brown E. G., Wood L. and Wood S., 1999. The Medical
Dictionary for Regulatory Activities (MedDRA). In
Drug Safety, Volume 20, Number 2, 109-117.
Erling O. and Mikhailov I., 2009. RDF support in the
Virtuso DBMS. In Studies in Computational
Intelligence, Volume 221/2009, 7-24.
Eschenbach A. von and Buetow K., 2006. Cancer
Informatic Vision: caBIG. In Cancer Informatics, vol
2. Pages 22-24.
Globus Toolkit from Globus Alliance. Available at:
http://www.globus.org/toolkit/ [16 July 2012]
HL7, Reference Information Model. Available at:
http://www.hl7.org/implement/standards/rim.cfm [16
July 2012]
INTEGRATE Driving Excellence in Integrative Cancer
Research. Available at: http://www.fp7-integrate.eu/
[16 July 2012]
Kazakov Y, Krötzsch M and Simancik F. 2011.
Concurrent classification of EL ontologies. In Lectures
Notes in Computer Science, Volume 7031/2011, 305-
320.
Kazakov Y., Krötzsch M. and Simancík F. 2012. ELK: a
reasoner for OWL EL ontologies. In Tech. rep.
Kiryakov A., Ognyanov D. and Manov D., 2005. OWLIM
– A pragmatic semantic repository for OWL. In
Lectures Notes in Computer Science, volume
3807/2005, 182-192.
Lawley M. and Bousquet C., 2010. Fast Classification in
Protege: Snorocket as an OWL2 EL Reasoner. In
Australasian Ontology Workshop.
Martin L., Anguita A., Graf N., Tsinakis M., Brochhausen
M., Rüping S., Sfakianakis S, Senqstaq T., Buffa F.
and Stenzhorn H., 2011. ACGT: advancing clinic-
genomic trials on cancer – four years of experience. In
Stud Health Technol Inform, 169:734-8.
McDonald C. J., Huff S. M., Suico J. G., Hill G., Leavelle
D., Aller R., Forrey A., Mercer K., DeMoor G., Hook
J., Williams W., Case J. and Maloney P., 2003.
LOINC, a universal standard for identifying laboratory
observations: a 5-year update. In Clinical Chemistry,
vol 49. No. 4, 624-633.
McGuinness D. L. and Van Harmelen F., 2004. OWL
Web Ontology Language Overview. W3C
Recommendation, available at: http://www.w3.org/
TR/owl-features/ [16 July 2012]
Murphy Shawn N, Weber G, Mendis M, Gainer V, Chueh
HC, Churchill S and Kohane I. 2010. Serving the
enterprise and beyond with informatics for integrating
biology and the bedside (i2b2). In J Am Med Inform
Assoc 2010; 17:124-130.
Neo-ALTTO, Neo-Adjunvant Lapatinib and Trastuzumab
Treatment Optimisation Trial. Available at:
http://www.alttotrials.com/neoaltto.php [16 July,
2012]
Parsia B. and Sirin E., 2004. Pellet: An OWL DL
reasoner. In ISWC 2004, 2004. ISWC.
Shearer R, Motik B and Horrocks I. 2008. HermiT: a
HEALTHINF2013-InternationalConferenceonHealthInformatics
40

highly-efficient OWL reasoner. In Proceedings of the
5th International Workshop on OWL: Experiences and
Directions (OWLED 2008): 26-27.
SNOMED Clinical Terms Core Content. Available at:
http://www.ihtsdo.org/snomed-ct/ [16 July 2012]
Stang P. E., Ryan P. B., Racoosin J. A., Overhage J. M.,
Hartzema AG, Reich C, Welebob E., Scarnecchia T
and Woodcock J., 2010. Advancing the science for
active surveillance: rationale and design for the
Observational Medical Outcomes Partnership. In Ann
Intern Med. 2010 Nov 2;153(9):600-6.
Tsarkov D. and Horrocks I., 2003. Reasoner prototype:
Implementing new reasoner with datatypes support. In
WonderWeb Project Deliverable.
TOP, Jules Bordet Institute. Topoisomerase II Alpha Gene
Amplification and Protein Overexpression Predicting
Efficacy of Epirubicin (TOP). Available at:
http://clinicaltrials.gov/ct2/show/NCT00162812 [16
July, 2012]
SemanticInteroperabilitySolutionforMulticentricBreastCancerTrialsattheIntegrateEUProject
41
