
A Channel Selection Method for EEG Classification based on
Exponentially Damped Sinusoidal Model
and Stochastic Relevance Analysis
Leonardo Duque Mu
˜
noz
1
, Carlos Guerrero-Mosquera
2
and German Castellanos-Dominguez
2
1
Grupo de Autom
´
atica y Electr
´
onica, Laboratorio MIRP, Instituto Tecnol
´
ogico Metropolitano, Medell
´
ın, Colombia
2
Departamento de Ingenier
´
ıa Electrica Electr
´
onica y Computaci
´
on, Universidad Nacional de Colombia,
Medell
´
ın, Colombia
Keywords:
Rhythms Decomposition, Seizure Detection, Feature Extractions, EEG Classification.
Abstract:
This work introduces a new methodology to select EEG channels related to epileptic seizures by electroen-
cephalogram (EEG) rhythms extraction. Rhythms extraction is an alternative to extract useful information
from specific band frequencies, analyze changes in the EEG signals, and detect brain abnormalities. In this
approach, the EEG signals are modeled by Exponentially Damped Sinusoidal model (EDS) and the EEG
rhythms extraction is based on Stochastic Relevance Analysis (SRA). Achieve results show that EDS model
combined with a stochastic relevance measure is a proper alternative for EEG classification of epileptic signals
and also could be used for EEG channel selection with seizure activity. The effectiveness of this approach is
compared in each experiment with other well known method for feature extraction called as Rhythmic Com-
ponent Extraction (RCE). This comparison was done based on the performance of the k-NN classifiers and
the channels selected were validated by visual inspection and topographic scalp map. The study uses real and
multi-channel EEG data and all the experiments have been supervised by an expert neurologist. We conclude
that the proposed scheme is a suitable approach for automatic seizure detection at a moderate computational
cost, also opening the possibility of formulating new criteria to select, classify or analyze abnormal EEGs
channels.
1 INTRODUCTION
Electroencephalographic signals (EEG) contain use-
ful information about the condition of the brain state
and their analysis is important to extract hidden infor-
mation or features that the human cannot directly de-
tect by visual inspection. Moreover, EEG signals with
epilepsy change continuously and to process every
EEG channel requires a long period of time, more vi-
sual analysis, high storage databases, and more com-
putational cost. To improve the efficiency in EEG
processing it is necessary to deal with multi-channel
processing and highlight the most pertinent features
of the signal. This allows to identify and extract EEG
rhythms faster, and also to see which parts of the brain
are the most affected for some abnormality.
Several authors have shown that epileptic seizures
could be decomposed into one or more components.
For example, the typical pattern or ictal rhythm in
mesial temporal lobe epilepsy appears as a high volt-
age blunt or sharp 5 − 7 Hz theta rhythm over one te-
mporal region (Nam et al., 2002). Other authors show
a principal epileptic track in band frequency range
between 2 − 7 Hz (Guerrero-Mosquera et al., 2010).
Therefore, EEG rhythms extraction could be used in
detecting brain abnormalities such as epilepsy dis-
ease.
There are different approaches propose in the liter-
ature for extracting EEG rhythms, such as wavelet de-
composition (Zalay et al., 2009), Independent Com-
ponent Analysis (ICA) (Sri and Rajapakse, 2008),
adaptive schema (Veluvolu et al., 2012), multi-
dimensional decomposition (Orekhova et al., 2011)
and frequency dominant characterization (Lodder and
Putten, 2011). Most of these studies are only fo-
cused on finding the onset and duration of the rhythm,
present many parameters to validate, show a depen-
dency with frequency resolution, but do not reflect the
true peak amplitude of the posterior dominant rhythm.
Other well know technique is the Rhythmic Com-
ponent Extraction (RCE), which combines multi-
channel signals with weights that are optimally sought
284
Duque Muñoz L., Guerrero-Mosquera C. and Castellanos-Dominguez G..
A Channel Selection Method for EEG Classification based on Exponentially Damped Sinusoidal Model and Stochastic Relevance Analysis.
DOI: 10.5220/0004196802840289
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2013), pages 284-289
ISBN: 978-989-8565-36-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

for such that the extracted component maximally con-
tains the power in the frequency range of interest and
suppresses that in unnecessary frequencies
1
. Other
methods proposed at the state-of-the-art need to vali-
date many parameters or demand computational cost
in training (Peters et al., 2001; Yang et al., 2012;
Duun-Henriksen et al., 2012).
In this paper, we introduce a new method for EEG
channel selection based on Exponentially Damped
Sinusoidal model (EDS) and Stochastic Relevance
Analysis (SRA). Our approach distinguishes variables
that represent effectively a “hidden” phenomena ac-
cording to stochastic variability measure, that is used
as relevance function (Sepulveda-Cano et al., 2011),
and detects the EEG channels with more seizure ac-
tivity. The effectiveness of our approach is compared
with RCE method both for EEG segment classifica-
tion problems and EEG channel selection. The classi-
fier used is the well known k-nearest neighbor (k-nn)
algorithm.
This paper is organized as follows: Section 2 in-
troduces the EDS model and the Stochastic Rele-
vance Analysis (SRA). Section 3 describes the EEG
databases and the experiments setup, and finally in
Section 4 and Section 5, the main results are discussed
and the principal conclusions with further work are
presented.
2 PROCESSING METHODS
The proposed method is based on two main steps:
(1) EEG is modeled by Exponentially Damped Si-
nusoidal model (EDS), and (2) EEG rhythms are
extracted by Stochastic Relevance Analysis (SRA).
Both methods are briefly explained as follows.
2.1 Exponentially Damped Sinusoidal
Model (EDS)
EDS model represents an EEG signal as a finite sum
of p discrete-time exponentially damped complex si-
nusoids:
x(n) =
p
∑
i=1
a
i
exp( j
ϕ
i
)exp((−d
i
+ j2
π
f
k
)n∆t) +
ε
(n)
n = 0, 1,...,N − 1
where n∆t is the time lapse between the origin and the
sample x(n), ∆t is the sampling interval, and
ε
(n) is
Gaussian white noise. The parameters of this model
are the amplitudes a
i
, the phases
ϕ
i
, the damping
1
More details of RCE algorithm in (Tanaka and Saito,
2008)
factors d
i
, and the frequencies f
i
. Since EDS are
a set of basis functions, they can be used to model
any arbitrary signal sufficiently closely, assuming that
the model order is high enough. Present study ad-
dresses the problem of estimating the model param-
eters when the EEG signal is embedded in noise by
using the subspace–based exponential data fitting pro-
posed in (De Clercq et al., 2005), where EEG signal
is stacked in Hankel data matrix. Then, the model pa-
rameters needed in Eq. (1) are estimated by Singular
Value Decomposition (SVD). More detail of EDS pa-
rameters estimation in (De Clercq et al., 2005).
2.2 Stochastic Relevance Analysis
(SRA)
Relevance analysis distinguishes variables that rep-
resente effectively the subjacent physiological phe-
nomena according to some evaluation measure. Such
representative variables are named relevant features,
whereas the evaluation measure is known as relevance
measure. Variable selection tries to reject those vari-
ables whose contribution to representation target is
none or negligible (irrelevant features), as well as
those that have repeated information (redundant fea-
tures). Thus, the first objective concerning the vari-
able selection stage is to define the concept of rele-
vance (Sepulveda-Cano et al., 2011). Let the set of
objects S
s
= {S
S
S
k
,k = 1,...,M} with M observations
described by a set of features s
i j
. In addition, each
sample is associated with one and only one element of
the set of class labels c
c
c =
{
c
(k)
∈ N : k = 1,··· ,K
}
,
where K is the number of classes to be considered.
Then, given S
s
, and for any feature s
i j
, the relevance
function
ρ
is defined as:
ρ
: R
1×T
−→ R
(S
s
,s
i j
) 7→
ρ
(S
s
,s
i j
) ∈ R (1)
where the relevant function
ρ
satisfies the following
properties (Sepulveda-Cano et al., 2011):
– Non-negativity,
ρ
(S
s
,s
i j
) ≥ 0, for all i.
– Nullity, the function
ρ
(S
s
,s
i j
) is null if feature s
i j
has not relevance at all.
– Non-redundancy, if s
′
i j
=
λ
s
i j
+
η
, where
the real-valued
λ
̸= 0, y
η
is some noyse
with zero mean and unit variance, then,
ρ
(S
s
,s
′
i j
) −
ρ
(S
s
,s
i j
)
→ 0
It is assumed that higher weights are associated with
the most relevant features. This work considers the
following unsupervised measure of relevance:
AChannelSelectionMethodforEEGClassificationbasedonExponentiallyDampedSinusoidalModelandStochastic
RelevanceAnalysis
285

c. Stochastic Variability: the time-varying relevance
measure is evaluated:
ρ
υ
(S
S
S
y
;
τ
) = [
χ
(1) ···
χ
(
τ
) ···
χ
(pT )]
⊤
, (2)
where
χ
(
τ
) = E {|
λ
2
j
v
j
(
τ
)|}, {
λ
j
: j = 1,...,q} is
the set of most relevant eigenvalues of the matrix
S
S
S
y
, and the scalar v
j
(
τ
) is the respective element
in the instant
τ
, and
τ
= 1,..., pT indexes each
of the relevance values calculated for the entire
set of time-varying data. To determine the rele-
vance related to each of the stochastic variables,
the Eq.(2) can be arranged to the relevance
matrix [
ρ
υ
1
(S
S
S
y
;t)···
ρ
υ
f
(S
S
S
y
;t)···
ρ
υ
F
(S
S
S
y
;t)]
⊤
,
where each row
ρ
υ
f
(S
S
S
y
;t) = [
χ
(( f − 1)T +
1)...
χ
(t)...
χ
( f T )] ∈ R
T ×1
shows the contribu-
tion of the s
s
s
ri
stochastic feature along fixed time
moments.
3 MATERIAL AND SETTING
This work uses two EEGs databases: one of them
(labeled as DB1), described in detail in (Guerrero-
Mosquera et al., 2010), consists in 6 adult epilep-
tic patients obtained in a restful wakefulness stage
and recorded at the Clinica Universitaria de Navarra,
Department of Neurophysiology (Pamplona, Spain).
All of them contained focal epileptiform activity,
according to experienced neurologists. We used 6
EEG recordings of 24 min length taken from 23-
th, 24-th and 25-th channels using the 10- 20 Inter-
national System of Electrode Placement with addi-
tional anterotemporal electrodes T1/T2. In practice,
raw EEG were filtered by a digital low-pass filter
with cut-off frequency of 20 Hz, sampled frequency
of 200 Hz and ocular artifacts rejected by Indepen-
dent Component Analysis (Guerrero-Mosquera and
Navia-Vazquez, 2012).
The other database (labeled as DB2) is a collec-
tion recorded at the Instituto de Epilepsia y Parkin-
son del Eje Cafetero (Pereira,Colombia). Each data
set from 35 patients contains 160 recorded scalp EEG
signals from 23-th, 24-th and 25-th channels corre-
sponding to electrodes placed on the head according
to the International 10-20 System of Electrode Place-
ment Standard. Set A holds 80 recordings labeled as
normal (seizure–free), whereas set E holds 80 record-
ings labeled as having epilepsy (epileptiform activity
by neurologist). Recordings have been sampled at a
frequency of 256 Hz, with 12 bits resolution and a 2-
min duration. All patients underwent clinical exami-
nation by neurologist. The data had been acquired in a
non–regulated conditions, and the noised data holds,
besides awake background EEG activity, the muscle
artifacts as well as 60 Hz power line interference.
3.1 Experimental Setup
Fig.1 summarizes the experimental outline proposed:
a) EEG is modeled by EDS and pre-processed follow-
ing (De Clercq et al., 2005); b) EEG rhythms are ob-
tained by Stochastic Relevance Analysis (SRA) and
RCE method; and c) Classifier performance and rele-
vant channels are obtained to determine the presence
of epilepsy. The selected channels will be evaluated
both visual inspection by expert neurologist and elec-
trode placement on topographic map.
Raw EEG
Modelling and
pre-processing:
EDS
Feature extraction:
RCE and
Stochastic analysis
k-NN
classifier
Classification
Channel
selection
Figure 1: A general scheme with the proposed method for
EEG classification and scalp localization of seizure activity.
EDS model uses four parameters: d determin-
ing which left singular vectors belong to the signal
subspace, N samples, and Hankel matrix dimension
L×M. The values used here corresponds to the values
suggested in (De Clercq et al., 2005) were N = 30720,
L = N/2 = 15371, M = L − 1 = 15370 and d = 0.01.
It was observed by visual inspection that this method
successfully had removed muscle artifact while the
epileptic activity had been enhanced.
In this work, we attempt to extract the frequency
band between 0.5 and 8 Hz, which is the most closely
related to epilepsy (Nam et al., 2002; Guerrero-
Mosquera et al., 2010). All experiments proposed
in this work use real and multi-channel EEG data
and have been supervised by an expert neurologist.
Regarding the relevance measure, both RCE and
stochastic variability are used to find the EEG chan-
nels with greater relevance weight in presence of
epileptic seizure and then finding approximately the
damaged brain region.
Each rhythm is used as a dynamic feature for the
classifier training. After obtaining the feature matrix,
Principal Component Analysis (PCA) is used as a fea-
ture extraction method to reduce the high dimension-
ality of the feature matrix. The number of principal
components (PCs) is selected based on the number of
PCs that maximizes the performance measures in the
classifier. Cross-validation procedure is used to eval-
uate the performance of proposed experiments, which
consists in dividing the database into 10 folds, each
one with an equal number of signals per class. A
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
286
k− nearest neighbors (k-nn) classifier is trained with
k = 5. The classification performance is measured by
means of the accuracy, sensitivity and specificity, de-
fined by:
A
cc
(%) =
N
c
N
T
∗ 100
Sens(%) =
N
T P
N
T P
+ N
FN
∗ 100;
Spec(%) =
N
T N
N
T N
+ N
FP
∗ 100;
where N
c
is the number of correctly classified pat-
terns, N
T
is the total number of patterns used to feed
the classifier, N
T P
is the number of true positives (ob-
jective class accurately classified), N
FN
is the number
of false negatives (objective class classified as refer-
ence class), N
T N
is the number of true negatives (ref-
erence class classified as objective class), and N
FP
is
the number of false positives (reference class classi-
fied as objective class).
Table 1: Accuracy values for each EEG channel by Stochas-
tic Relevance Analysis (SRA) for all patients (DB1). Values
in bold type are those selected by neurologist through a vi-
sual inspection. Note the correspondence between high val-
ues obtained by SRA and EEG channels chosen by visual
inspection.
EEG
Patient
EEG
Patient
EEG
Patient
1 2 3 4 5 6
F4 0.16 0.06 Fp1 0.19 F4 0.59 0.27 0.66
FP2 0.46 0.06 F3 0.08 Fp2 0.14 0.13 0.49
F3 0.43 0.20 C3 0.11 F3 0.32 0.11 0.40
FP1 0.18 0.36 P3 0.05 Fp1 0.18 0.15 0.41
T6 0.31 0.09 O1 0.04 T6 0.50 0.24 0.31
T5 0.98 0.15 F7 0.06 T5 0.20 0.14 0.36
O2 0.48 0.45 T3 0.36 O2 0.11 0.01 0.29
O1 0.62 0.31 T5 0.19 O1 0.26 0.07 0.20
F7 0.82 0.32 Fp2 0.17 F7 0.77 0.11 0.39
F8 0.58 1.00 F4 0.11 F8 0.25 0.48 0.74
T3 0.98 0.15 C4 0.46 T3 0.13 0.70 0.32
T4 0.22 0.75 P4 0.18 T4 0.89 0.05 0.84
C4 0.51 0.47 O2 0.16 C4 0.20 0.03 0.20
C3 0.35 0.15 F8 0.32 C3 0.21 0.06 0.21
P4 0.75 0.16 T4 1.00 P4 0.27 0.23 0.25
P3 0.59 0.12 T6 0.88 P3 0.07 0.15 0.29
Cz 0.77 0.42 Fz 0.17 Cz 0.69 0.14 0.15
Ecgp 0.11 0.01 Cz 0.15 Ecgp 0.6 0.01 0.08
Pz 0.85 0.59 Pz 0.13 Pz 0.40 0.18 0.38
T1 1.00 0.15 A1 0.10 T1 1.00 1.00 0.13
T2 0.29 0.63 A2 0.72 T2 0.08 0.09 1.00
Fz 0.47 0.53 Ecg1 0.02 A1 0.84 0.91 0.59
Ecgn 0.04 0.03 T1 0.21 A2 0.34 0.12 0.82
T2 0.13 Fz 0.25 0.22 0.62
Ecgn 0.01 0.01 0.07
4 RESULTS
This section shows the effectiveness of the methodol-
ogy proposed by comparing the approach with Rhyth-
mic Component Extraction (RCE) in two sceneries:
channel selection and EEG classification.
4.1 Channel Selection
Table 1 shows the estimated stochastic relevance val-
ues for each EEG channel (DB1 data), computed by
SRA. Values in bold type are those selected by neu-
rologist through a visual inspection. Note the corre-
spondence between high values obtained by SRA and
EEG channels chosen by visual inspection.
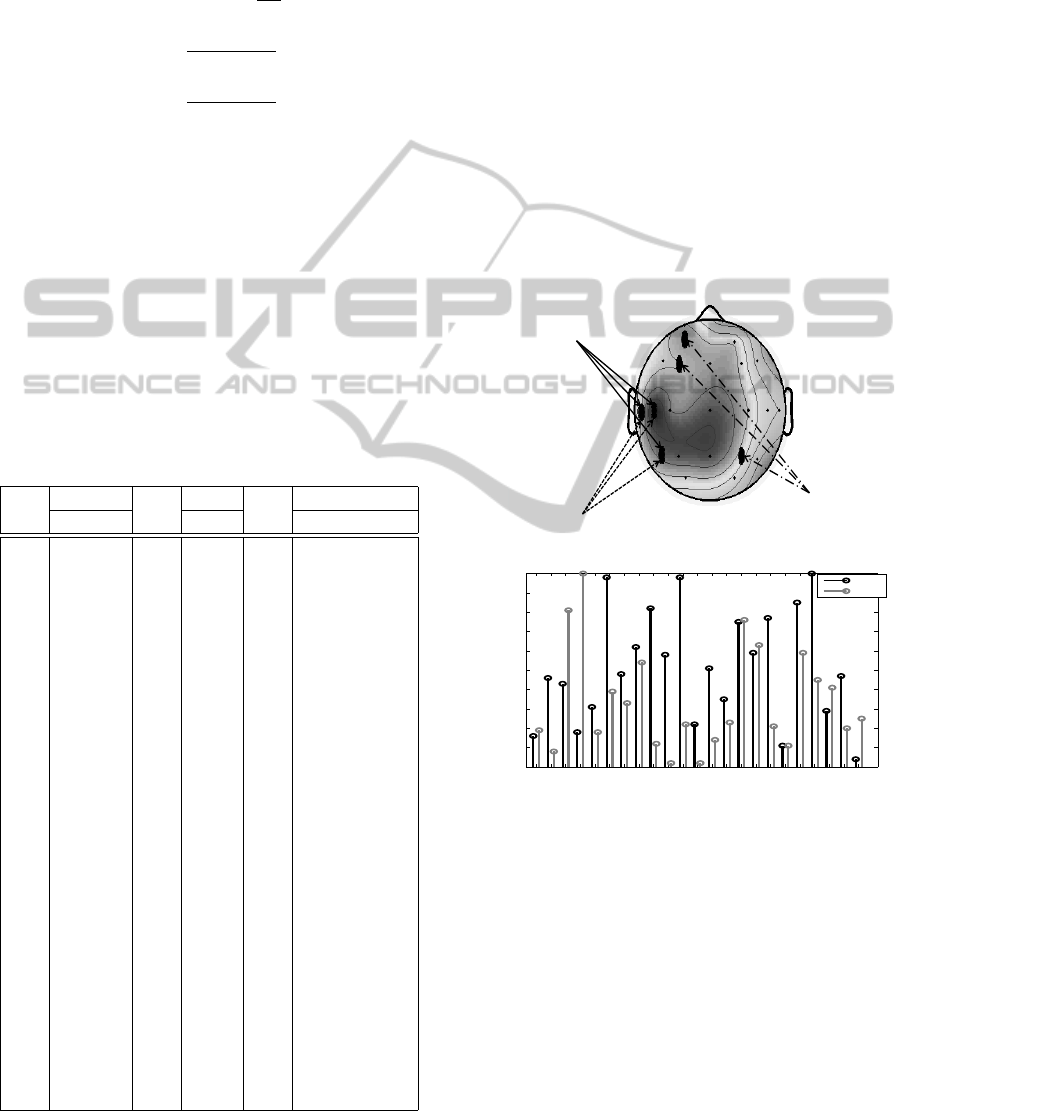
Localization of channels with high relevance values in single EEG data
(Pacient 1)
Channels by visual inspection
(T1, T3 and T5)
Channels by stochastic relevance
(T1, T3 and T5)
Channels by RCE
(FP1,F3 and P4)
F4F´2F3F´1T6 T5O2O1F7F8T3T4C4C3P4 P3CzEpPz T1T2 Fz En
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
EEG Channels
Normalized Amplitude w
SRA
RCE
Figure 2: Comparison of the channels selected by RCE,
stochastic relevance (SRA) and visual inspection at patient
1 (DB1 database). Upper: Localization of the channels se-
lected on scalp topographic map. Black areas represent a
highest energy concentration than grey areas. Bottom: Rel-
evance values obtained by RCE method and stochastic rel-
evance for each channel. Note that EEG channels with high
weighting by stochastic relevance (bottom) correspond to
channels located on epileptogenic region (upper).
Fig. 2 (upper) shows corresponding channels selected
by RCE and stochastic relevance (SRA) with the vi-
sual inspection on scalp topographic map at patient
1 (DB1). It can be seen that high values achieved by
RCE method do not correspond to channels chosen by
the neurologist through visual inspection. The Fig. 2
AChannelSelectionMethodforEEGClassificationbasedonExponentiallyDampedSinusoidalModelandStochastic
RelevanceAnalysis
287

(bottom) depicts relevance values obtained for each
DB1 channel at the patient 1 by RCE method and
stochastic relevance. Note that SRA measures suc-
cessfully selects the channels with more seizure ac-
tivity (T1, T3 and T5) showing high relevance values
close to 1. All results show that our method provide
a better medical interpretability about epileptic region
compared to the weights computed by RCE method.
Shortest weights values corresponding to EEG deriva-
tions are discarded in this figure.
4.2 EEG Segment Classification
Table 2 shows classification results achieved in DB1
and DB2 databases by RCE and stochastic variability
(SRA). As seen from Table 2, there is an improve-
ment in terms of classifier performance when we use
stochastic relevance, DB1 achieves 95.38% and DB2
achieves 94.60% in accuracy values, compare to RCE
that present accuracy values for DB1 (93.60%) and
DB2 (92.10%).
Table 2: Comparison in classification performance of RCE
and SRA by method by k-NN algorithm.
Method DB Acc( %) Sens(%) Spec(%)
RCE
DB1 93.60 93.60 93.60
DB2 92.10 93.85 90.35
SRA
DB1 95.38 95.90 96.23
DB2 94.60 94.60 94.32
Note that values in classifier performance are
close each to other, even though DB2 is more con-
taminated by artifacts than DB1, which has a pre-
processing to eliminate ocular movements (Guerrero-
Mosquera and Navia-Vazquez, 2012). The proposed
model is then stable for EEG classification problems
in presence of noise.
5 CONCLUSIONS AND FUTURE
WORK
A method for classification and channel selection for
EEG multi-channel data with epilepsy is proposed.
This method, based on Exponentially Damped Sinu-
soidal model (EDS) and Stochastic Relevance Anal-
ysis (SRA), is simple and do not requires high com-
putational cost in training than others methods pro-
posed at the state-of-the-art (Peters et al., 2001; Yang
et al., 2012; Duun-Henriksen et al., 2012). Achieved
results show this method is an alternative for extract-
ing relevant EEG rhythms and selecting EEG chan-
nels with epileptic activity. Results validated by ex-
perts through visual inspection and scalp topographic
map, show that the approach also provides a better
medical support in epileptic region localization.
Future work includes: comparing our approach with
other seizure detection methods proposed in the state-
of-the-art; exploring other brain abnormalities such
as Alzheimer, sleep disorders and dementia; exploit-
ing the features for epileptogenic region analysis, and
consider the feasibility of our method to seizure an-
ticipation.
ACKNOWLEDGEMENTS
This research is carried out under the grant Centro de
Investigaci
´
on e Innovaci
´
on de Excelencia ARTICA,
sponsored by COLCIENCIAS and Convocatoria de
apoyo a tesis de posgrado DIMA 2011, Universidad
Nacional de Colombia, project 13753. Also thanks
to Instituto de Epilepsia y Parkinson del Eje Cafetero
(Pereira,Colombia) with the EEG data collection.
REFERENCES
De Clercq, W., Vanrumste, B., Papy, J.-M., Van Paesschen,
W., and Van Huffel, S. (2005). Modeling common
dynamics in multichannel signals with applications
to artifact and background removal in EEG record-
ings. IEEE Transactions on Biomedical Engineering,
52(12):2006–2015.
Duun-Henriksen, J., Kjaer, T. W., and et al (2012). Chan-
nel selection for automatic seizure detection. Clinical
Neurophysiology, 123:84–92.
Guerrero-Mosquera, C., Malanda-Trigueros, A., Iriarte-
Franco, J., and Navia-Vazquez, A. (2010). New fea-
ture extraction approach for epileptic EEG signal de-
tection using time-frequency distributions. Med. Biol.
Eng. Comput., 48:321–330.
Guerrero-Mosquera, C. and Navia-Vazquez, A. (2012). Au-
tomatic removal of ocular artifacts using Adaptive Fil-
tering and Independent Component Analysis for EEG
data. IET Signal Processing, 6:99–106.
Lodder, S. and Putten, M. V. (2011). Automated eeg anal-
ysis: Characterizing the posterior dominant rhythm.
Journal of Neuroscience Methods , 200:86–93.
Nam, H., Yim, T.-G., Han, S. K., Oh, J.-B., and Lee,
S. K. (2002). Independent Component Analysis of ic-
tal EEG in medial temporal lobe epilepsy. Epilepsia,
43:160–164.
Orekhova, E., Elam, M., and Orekhov, V. (2011). Adaptive
estimation of EEG-rhythms for optimal band identi-
fication in BCI. Journal of Neuroscience Methods,
195:47–60.
Peters, B., Pfurtscheller, G., and Flyvbjerg, H. (2001). Au-
tomatic differentiation of Multichannel EEG signals.
IEE Trans. On Bio Med Eng, 48:111–116.
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
288

Sepulveda-Cano, L., Acosta-Medina, C., and Castellanos-
Dominguez, G. (2011). Relevance Analysis of
Stochastic Biosignals for Identification of Patholo-
gies. EURASIP Journal on Advances in Signal Pro-
cessing, 2011:10.
Sri, K. S. and Rajapakse, J. C. (2008). Extracting EEG
rhythms using ICA-R. IEEE International Joint Con-
ference on Neural Networks, pages 2133–2138.
Tanaka, T. and Saito, Y. (2008). Rhythmic component ex-
traction for multi-channel eeg data analysis. In Acous-
tics, Speech and Signal Processing, 2008. ICASSP
2008. IEEE International Conference on, pages 425–
428.
Veluvolu, K., Wang, Y., and Kavuri, S. (2012). Adaptive
estimation of EEG-rhythms for optimal band identi-
fication in BCI. Journal of Neuroscience Methods,
212:163–172.
Yang, J., Singh, H., and et al. (2012). Channel selection and
classification of electroencephalogram signals: An ar-
tificial neural network and genetic algorithm-based
approach. Artificial Intelligence in Medicine, 55:117–
126.
Zalay, O., Kang, E., Cotic, M., Carlen, P., and Bardakjian,
B. (2009). A wavelet packet-based algorithm for the
extraction of neural rhythms. Annals of Biomed. Eng.,
37:595–613.
AChannelSelectionMethodforEEGClassificationbasedonExponentiallyDampedSinusoidalModelandStochastic
RelevanceAnalysis
289
