
A Literature Evaluation of CUDA Compatible Sequence Aligners
Yang Liu, Jiang-Yu Li, Yi-Qing Mao, Xiao-Lei Wang and Dong-Sheng Zhao
Institute of Health Service and Medical Information, Academy of Military Medical Sciences, Taiping Road, Beijing, China
Keywords: CUDA, Sequence Aligner, Performance per Watt, Price-performance, Programming Complexity.
Abstract: The rapidly accumulating biological data generated by next-generation sequencer motivate the development
of improved tools for sequence alignment. Many technologies have been proposed for this purpose, and one
of them is GPU computing. Existing acceleration of sequence aligners using GPU computing
overemphasize speed. However, other factors such as accuracy, performance per watt, price-performance
and programming complexity are also important and need to be considered. Based on the existing literatures
of GPU-based sequence aligners, this paper gives a literature evaluation of these sequence aligners from the
above perspectives, in order to determine the usability of the tremendous GPU-based sequence aligners.
1 INTRODUCTION
With the coming of big data era, traditional widely
adopted sequence aligners such as BLAST can no
longer meet the need to analyse the rapidly
accumulating sequence data. In response to this
challenge, new algorithms have been proposed to
increase speed with no sacrifice in sensitivity or
accuracy. One example is USEARCH (Edgar, 2010).
Some new technologies such as GPU computing
have also been used to accelerate alignment. Several
traditional sequence aligners have been implemented
with CUDA. However, to highlight performance the
authors tend to overemphasize the gain of speed.
Besides speed, Accuracy, performance per watt,
price-performance and programming complexity are
also important factors that need to be concerned, yet
omitted by almost all of the authors. Should I move
my task to the GPU platform? This is the question
that this article will answer. This article aims to
systematically evaluate the usability of these
tremendous aligners from a comprehensive point of
view.
2 MATERIALS AND METHODS
2.1 Materials
Several sequence aligners have been migrated to
GPU. We investigate the widely-known CUDA
compatible sequence aligners that were developed in
recent years through the method of literature search.
The hardware and speedup information are listed in
Table 1. The speed performances of some aligners
vary with dataset, so we calculated the average
speedup if data is provided, otherwise the mentioned
speedup is used. Some aligners such as NCBI
BLAST and BWA can exploit the multicore feature
of CPU, so we use the speedup of the GPU-based
aligner compared with its multi-threaded counterpart
in later calculations under this situation. The number
of threads is equal to the number of CPU cores.
2.2 Methods
Most GPU-based sequence aligners achieve some
extent of speedup compared with the corresponding
aligners implemented on CPU. GPU computing is a
solution for high performance computing and has
much potential, but it is not a panacea. There are
some cases where GPU computing may not be
beneficial. In this section, we discuss four major
factors regarding the use of GPU: accuracy,
performance per watt, price-performance and
programming complexity.
Some of the CUDA compatible aligners don’t
discuss the accuracy in the papers. We can infer
from the implementation of algorithm.
Performance per watt of these aligners compared
with their corresponding CPU-based aligners is
evaluated by PPW
GPU+CPU
/PPW
CPU
. It is calculated
with the following formula.
268
Liu Y., Li J., Mao Y., Wang X. and Zhao D..
A Literature Evaluation of CUDA Compatible Sequence Aligners .
DOI: 10.5220/0004191202680271
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 268-271
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
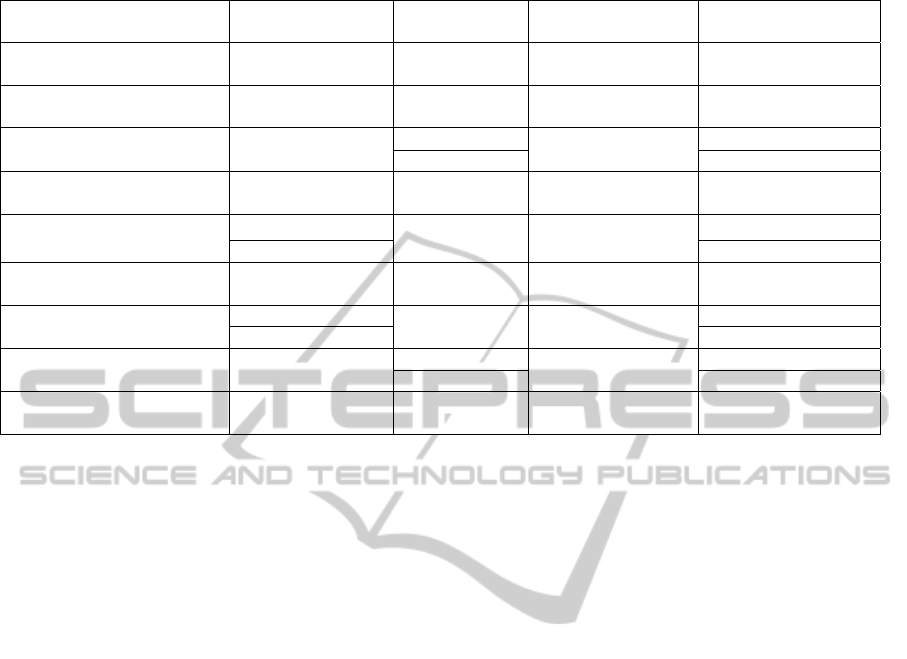
Table 1: Information about GPU-based sequence aligners and comparison against their CPU-based counterparts.
GPU-based
Aligner
GPU
CPU-based
Aligner
CPU Speedup
MUMmerGPU
(Schatz et al., 2007)
GeForce 8800 GTX MUMmer
Intel Xeon 5160
(dual-core)
3.47x-3.79x
GPU accelerated HMMER
(Walters et al., 2008)
GeForce 8800 GTX HMMER
2.0 GHz Intel Xeon
(quad-core)
18.30x
SW-CUDA
(Manavski and Valle, 2008)
GeForce 8800 GTX
SSEARCH
3 GHz Pentium 4
(single-core)
15.30x
NCBI BLAST 0.94x
GSW
(Striemer and Akoglu, 2009)
Tesla C870 SSEARCH
3.2 GHz Pentium 4
(single-core)
8.50x
CUDASW++
(Liu et al., 2009)
GeForce GTX 280
NCBI BLAST
Intel Xeon 3.0 GHz
(dual-core)
3.48x(BLOSUM50)
GeForce GTX 295 5.31x(BLOSUM50)
GPU-BLAST
(Vouzis and Sahinidis, 2011)
Tesla C2050 NCBI BLASTP
Intel Xeon 2.67GHz
(six-core)
1.59x
CUDA-BLASTP
(Liu et al., 2011)
GeForce GTX 280
NCBI BLASTP
Intel i7-920
(quad-core)
1.90x
GeForce GTX 295 2.82x
SOAP3
(Liu et al., 2012)
GeForce GTX 580
BWA
3.07 GHz
(quad-core)
2.84x
Bowtie 5.60x(3 mismatches)
BarraCUDA
(Klus et al., 2012)
Tesla M2090 BWA
Intel Xeon X5670
(six-core)
1.09x
PPW
GPU+CPU
/PPW
CPU
= S*P
CPU
/(P
GPU
+P
CPU
) (1)
P
GPU
and P
CPU
stand for the power dissipation of
GPU and CPU respectively. S stands for speedup.
Note that PPW
GPU+CPU
/PPW
CPU
is used because GPU
computing is a heterogeneous architecture and the
GPU-based aligners cannot run without CPU.
Price-performance is evaluated by PPR
GPU+
Sys
/PPR
Sys
, where PPR stands for Price/Performance
Ratio. It is calculated with the following formula.
PPR
GPU+S
y
s
/PPR
S
y
s
= (Pr
GPU
+Pr
S
y
s
)/(Pr
S
y
s*
S) (2)
Pr
GPU
and Pr
Sys
are the price of GPU and system
respectively. S stands for speedup. Since different
pcs and servers are used, to be even we use a range
(from $1000 to $3000) to denote the price of system.
The price of a PC and a server are about $1000 and
$3000 respectively.
There is no straightforward criterion to evaluate
programming complexity. But some factors such as
degree of popularization can be a point of view
which indicates the complexity.
3 RESULTS
3.1 Accuracy
In original literature, only three papers (CUDA-
BLASTP, SOAP3 and BarraCUDA) give detailed
discussion of accuracy. SOAP3 and BarraCUDA get
nearly the same accuracy with their corresponding
CPU-based aligners. Although CUDA-BLASTP gets
a little worse result when aligning sequences shorter
than 128, it is much faster (CUDA-BLASTP
achieves speedup of up to 10.0 compared with
sequential NCBI BLASTP). Other aligners get the
same accuracy with their corresponding CPU-based
aligners.
3.2 Performance per Watt
GPU computing is energy-effective for many
applications. We gather the power dissipation
information in Table 2 according to the hardware
information listed in Table 1.
Higher PPW
GPU+CPU
/PPW
CPU
value indicates
better performance per watt. If PPW
GPU+CPU
/PPW
CPU
is higher than 1, it means the GPU-based aligner is
more energy-effective.
Six out of nine get PPW
GPU+CPU
/PPW
CPU
value
higher than 1. These aligners can be divided into
different groups. SOAP3 and BarraCUDA are short-
read aligners. CPU is a better choice for
BarraCUDA. SW-CUDA, GSW and CUDASW++
are implementations of Smith-Waterman algorithm
on GPU. They generally get a higher PPW
GPU+CPU
/
PPW
CPU
compared with GPU-BLAST and CUDA-
BLASTP. This is because Smith-Waterman
algorithm can better utilize the parallelism of GPU
compared with NCBI BLAST. Sequence alignment
with a suffix tree such as MUMmer might be
expected to be a poor candidate for GPU, but
MUMmerGPU stills get a relative good result
because MUMmer cannot exploit the multicore
feature of modern CPU.
ALiteratureEvaluationofCUDACompatibleSequenceAligners
269

Table 2: Power dissipations of hardware used in the papers. The data are from (Intel, 2012), (AMD, 2012), (Nvidia Tesla,
2012) and (geeks3d, 2012). The asterisk indicates the range of power dissipation since the model of CPU is uncertain.
GPU-based
Aligner
Power dissipation
of GPU (watts)
CPU-based
Aligner
Power dissipation
of CPU (watts)
Speedup
PPW
GPU+CPU
/PPW
CPU
MUMmerGPU 145 MUMmer 80 3.47x-3.79x 1.23-1.35
GPU accelerated
HMMER
145 HMMER 80 18.30x 6.51
SW-CUDA 145
SSEARCH
81.9-89*
15.30x 5.52-5.82
NCBI-BLAST 0.94x 0.34-0.36
GSW 170.9 SSEARCH 82-86* 8.50x 2.76-2.85
CUDASW++
236
NCBI BLAST 80-95*
3.48x(BLOSUM50) 0.88-1.00
289 5.31x(BLOSUM50) 1.15-1.31
GPU-BLAST 238 NCBI BLASTP 130 1.59x 0.56
CUDA-BLASTP
236
NCBI BLASTP 130
1.90x 0.67
289 2.82x 0.87
SOAP3 244
BWA
95-130*
2.84x 2.1-2.6
Bowtie 5.60x (3 mismatches) 1.57-1.94
BarraCUDA 225 BWA 95 1.09x 0.32
Table 3: Prices of the hardware. The data are from the paper (Schatz et al., 2007), (Amazon, 2012) and (Tweakers, 2012).
M denotes the price of system and ranges from $1000 to $3000.
GPU-based
Aligner
Price of
GPU ($)
CPU-based
Aligner
Price of
System($)
Speedup
PPR
GPU+Sys
/PPR
S
y
s
MUMmerGPU 529 MUMmer 882+M 3.47x-3.79x 0.30-0.37
GPU accelerated
HMMER
699 HMMER 389+M 18.30x 0.07-0.08
SW-CUDA 681
SSEARCH
106+M
15.30x 0.08-0.11
NCBI BLAST 0.94x 1.30-1.72
GSW 1400 SSEARCH 93+M 8.50x 0.17-0.27
CUDASW++
426
NCBI BLAST 200+M
3.48x(BLOSUM50) 0.33-0.39
559 5.31x(BLOSUM50) 0.22-0.28
GPU-BLAST 2781 NCBI BLASTP 1252+M 1.59x 1.04-1.41
CUDA-BLASTP
439
NCBI BLASTP 343+M
1.90x 0.60-0.70
549 2.82x 0.41-0.50
SOAP3 648
BWA
259+M
2.84x 0.42-0.53
Bowtie 5.60x (3 mismatches) 0.21-0.27
BarraCUDA 2400 BWA 1684+M 1.09x 1.39-1.74
3.3 Price-performance
For large research institutes, cost is not the
bottleneck at most times, but it is still an important
factor. Table 3 lists the prices of the hardware. Some
GPUs are out of production now, and the price of
hardware changes with time. So we use the retail
prices when the paper is published.
If PPR
GPU+Sys
/PPR
Sys
value is lower than 1, it
means the GPU-based aligner is more economical
than the corresponding aligner based on CPU.
Seven out of nine get PPR
GPU+Sys
/PPR
Sys
value lower
than 1. The different groups of sequence aligners
give similar result to those of energy efficiency
performance. The GPU-based aligners are generally
more economical than CPU-based aligners except
GPU-BLAST and BarraCUDA.
3.4 Programming Complexity
We first consider the emerging time of some GPU-
based aligners and their corresponding aligners
based on CPU. SOAP3 and BarraCUDA are
released in 2012, and their corresponding CPU-
based aligners SOAP2 and BWA are released in
2009. The intervals are both three years which are
much longer compared with the transplant interval
of cloud-based applications. On the other hand,
Wikipedia (Sequence alignment software, 2012)
provides a list of sequence aligners, but only a small
portion of them are implemented with GPU.
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
270

4 DISCUSSIONS
From the user’s standpoint, to obtain optimal result
accuracy should always be preferred to speed. Most
of the sequence aligners mentioned above complies
with this principle.
From above result we can see that even though
some sequence alignment algorithms such as
BLAST and MUMmer are not intrinsically suitable
for parallelization, they still get considerable
speedup without loss of accuracy. At the same time,
the performance per watt and price-performance of
GPU is better for most of the sequence aligners.
GPU computing is still a low-cost and energy-
efficient solution for high performance computing.
The programming complexity of CUDA slows
down the popularization of GPU computing in some
extent. But with the release of new NVIDIA GPU
compute architecture and the spread of some parallel
computing standards such as OpenACC (OpenACC,
2012) and OpenHMPP (OpenHMPP, 2012), GPU
has arguably become as easy, if not easier, to
program than multicore CPUs.
From the four factors discussed above we can see
that GPU computing is a sound choice for sequence
alignment. But there are more issues you may care
about. First, we can see that the existing GPU-based
sequence aligners are far from exploiting the
computation capability of GPU, though accelerate
the alignment to some extent. Second, further
development is needed for the usability of GPU-
based aligners. In the result, CUDASW++ is faster
and more accurate than NCBI BLAST. So why not
to choose CUDASW++? Usability is an important
factor that influences the user’s choice. The GPU-
based aligners are mainly developed for academic
research, most of which lacks later maintenance and
upgrade. The features of these GPU-based aligners
are far less than that of CPU-based aligners. The
solution of usability calls for more professional
programmers and algorithm designers to help with
the research of bioinformatics.
REFERENCES
Amazon, [online], Available: http://www.amazon.com
Accessed 2012-07-12.
AMD official website, [online], Available: http://products.
amd.com/pages/opteroncpuresult.aspx?AspxAutoDete
ctCookieSupport=1 Accessed 2012-10-20.
Edgar, R. C., 2010. Search and clustering orders of
magnitude faster than BLAST. Bioinformatics, vol. 26,
no. 19, pp. 2460-2461.
Geeks3d, [online], Available: http://www.geeks3d.com/20
090618/graphics-cards-thermal-design-power-tdp-data
base Accessed 2012-07-12.
Intel official website, [online], Available: http://ark.intel.c
om Accessed 2012-10-20.
Klus, P., Lam, S., Lyberg, D., Cheung, M. S., Pullan, G.,
McFarlane, I., Yeo, G., Lam, B., 2012. BarraCUDA -
a fast short read sequence aligner using graphics
processing units. BMC Research Notes, vol. 5, no. 1,
pp. 27.
Liu, Y., Maskell, D. L., Schmidt, B., 2009. CUDASW++:
optimizing Smith-Waterman sequence database
searches for CUDA-enabled graphics processing units.
BMC Res Notes, vol. 2, pp. 73.
Liu, W., Schmidt, B., Muller-Wittig, W., 2011. CUDA-
BLASTP: accelerating BLASTP on CUDA-enabled
graphics hardware. IEEE/ACM Transactions on
Computational Biology and Bioinformatics (TCBB),
vol. 8, no. 6, pp. 1678-1684.
Liu, C. M., Wong, T., Wu, E., Luo, R., Yiu, S. M., Li, Y.,
Wang, B., Yu, C., Chu, X., Zhao, K., Li, R., Lam, T.
W., 2012. SOAP3: Ultra-fast GPU-based parallel
alignment tool for short reads. Bioinformatics, doi:
10.1093/bioinformatics/bts061.
Manavski, S. A., Valle, G., 2008. CUDA compatible GPU
cards as efficient hardware accelerators for Smith-
Waterman sequence alignment. BMC Bioinformatics,
vol. 9, no. Suppl 2, pp. S10.
Nvidia Tesla, [online], Available: http://en.wikipedia.org/
wiki/Nvidia_Tesla Accessed 2012-07-12.
OpenACC, [online], Available: http://www.openacc-stand
ard.org Accessed 2012-07-12.
OpenHMPP, [online], Available: http://www.openhmpp.or
g Accessed 2012-07-12.
Schatz, M. C., Trapnell, C., Delcher, A. L., Varshney, A.,
2007. High-throughput sequence alignment using
Graphics Processing Units. BMC Bioinformatics, vol.
8, pp. 474.
Sequence alignment software, [online], Available:
http://en.wikipedia.org/wiki/List_of_sequence_alignm
ent_software Accessed 2012-07-12.
Striemer, G. M., Akoglu, A., 2009. Sequence Alignment
with GPU: Performance and Design Challenges. In
IEEE International Symposium on Parallel and
Distributed Processing.
Vouzis, P. D., Sahinidis, N. V., 2011. GPU-BLAST: using
graphics processors to accelerate protein sequence
alignment. Bioinformatics, vol. 27, no. 2, pp. 182-188.
Walters, J. P., Balu, V., Kompalli, S., Chaudhary, V., 2008.
Evaluating the use of GPUs for life science
applications. Buffalo, NY: The State University of
New York, http://www.cse.buffalo.edu/tech-
reports/2008-10.pdf. Accessed 2012-03-19.
Tweakers, [online], Available: http://tweakers.net/pricewat
ch Accessed 2012-10-20.
ALiteratureEvaluationofCUDACompatibleSequenceAligners
271
