
An Ontology-based Framework for Syndromic Surveillance Method
Selection
Gabriela Henriques and Deborah Stacey
School of Computer Science, University of Guelph, 50 Stone Road East, Guelph, Canada
Keywords:
Ontology, Syndromic Surveillance, Algorithms, Optimization, Leveraging Knowledge.
Abstract:
Syndromic surveillance is the detection of a disease outbreak or bioterrorist attack. The process of surveil-
lance includes various steps: data collection, data analysis and result interpretation. The goal of syndromic
surveillance is to be able to make a rapid and accurate diagnostic of a potential outbreak. Method types range
from traditional statistical approaches to algorithms which have been adapted from other fields. With a variety
of options it can be difficult selecting the method best suited for analysis on a given set of data. This paper will
focus on developing an ontology-based framework for selecting the best suited method(s) for data analysis,
focusing on the end-users perspective.
1 INTRODUCTION
Public health surveillance is the monitoring of bioter-
rorist attacks and disease outbreaks (Henning, 2004;
McDade and Franz, 1998). The term syndromic is
commonly used when discussing surveillance to em-
phasize its focus on early detection of an attack or out-
break in a geographical location. In the past decade,
the need for syndromic surveillance has become pri-
oritized; the early detection of an outbreak can pre-
vent massive illness and death (Henning, 2004; Mc-
Dade and Franz, 1998).
An ontology is a form of knowledge representa-
tion, it is used to represent a set of concepts and their
relationships within a domain. An ontology has the
ability to reason with the entities of a domain, and
can thus be used to describe the domain itself. Many
ontology-based frameworks have been developed in
various application areas to aid in data collection, or-
ganization, and classification.
A variety of methods exist that can be used for
data analysis when determining whether a potential
outbreak has occurred within a region. Some of the
approaches include benchmark methods such as cu-
mulative sum, and moving average. Other meth-
ods include more non-traditional approaches that have
been adapted from different fields, such as neural
networks and genetic algorithms. Many syndromic
surveillance systems incorporate a variety of meth-
ods in their program, providing the end-user (analyst)
with different options to use for analysis. However,
these systems all have a different set of requirements
which may not be best suited for the technology cur-
rently used by the user. As well, the methods admin-
istered in a system may not be the most appropriate
for a set of data that needs to be analyzed.
This paper will start off by providing background
information on syndromic surveillance and existing
systems in section 2. Section 3 will provide a motive
and proposal for an ontology-based framework to aid
in the selection of a set of methods most appropriate
for a given set of data, focussing on the requirements
specified by an end-user. Section 4 will discuss two
disease detection examples, analyzing important pa-
rameters to be considered for the proposed system.
The paper will end off with future work directions
provided in section 5.
2 BACKGROUND
2.1 Syndromic Surveillance
Surveillance relies on three main steps: data collec-
tion, data analysis and result interpretation (Buck-
eridge et al., 2008). Data collection involves the gath-
ering of data from a variety of sources including hos-
pital emergency department (ED) records, over-the-
counter (OTC) pharmaceutical sales, and news reports
(Buckeridge et al., 2002; Lu et al., 2008; Crubezy
et al., 2005). In more recent years, the collection pro-
396
Henriques G. and Stacey D..
An Ontology-based Framework for Syndromic Surveillance Method Selection.
DOI: 10.5220/0004146003960400
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2012), pages 396-400
ISBN: 978-989-8565-30-3
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

cess has evolved from a time-consuming data gather-
ing process, to automatic real-time data collected and
distributed for analysis. Data analysis depends on the
process of data collection. In order for analysis to be
effective, a variety of methods should be considered
so that the best suited technique is selected.
Two key factors which must be taken into consid-
ered when discussing data collection for surveillance
are: timeliness and specificity (Buckeridge et al.,
2008). The timeliness of outbreak detection is very
important aspect in syndromic surveillance. A one-
day delay in detection could result in a loss of millions
of dollars, and massive illness and death(Buckeridge
et al., 2002). At present, new systems have automated
the process of gathering data in order to aid in the
speed of collection; public health departments now
have access to real-time data sets coming from a vari-
ety of different sources (Buckeridge et al., 2002; Tsui
et al., 2003).
Another factor that must be taken into considera-
tion when dealing with data collection, is the speci-
ficity of the data. There are various sources from
which syndromic surveillance data is collected, some
of the forms of data include emergency department di-
agnostics, over-the-counter pharmaceutical sales, and
news reports (Buckeridge et al., 2002; Lu et al., 2008;
Crubezy et al., 2005). Generally, data can be grouped
into three different categories of sources: pre-clinical,
clinical pre-diagnostic and diagnostic (Buckeridge
et al., 2002). Pre-clinical data is gathered before go-
ing to a health care centre. This information typically
consists of school or office absenteeism and is not
very specific. Clinical pre-diagnostic includes infor-
mation such as test orders, signs, symptoms and over-
the-counter sales. This information is timely, and rel-
atively specific. Diagnostics are data gathered from
test results and case interviews; these forms of data
are very specific however they are not timely. In or-
der for analysis methods to be accurate and effective,
it is important that the data is specific. However, due
to the need for timely detection, it has become more
popular to analyze clinical pre-diagnostic information
(Buckeridge et al., 2008). Since this data incurs a loss
of specificity during the collection process, some of
the algorithms used for detection may be ineffective
without taking extra precautions on how to interpret
the data in a classified manner.
2.2 Syndromic Surveillance Systems
Vast amounts of data are gathered in syndromic
surveillance. In order to perform a rapid and accurate
analysis, the most competent method must be used.
Aberration-detection algorithms are commonly used
for data analysis. These algorithms include statisti-
cal benchmark methods such as cumulative sums, re-
gression models, moving average calculations etc ...
along with knowledge-based algorithms such as ar-
tificial neural networks, genetic algorithms and on-
tologies. Some of these algorithms were developed
specifically for surveillance, while others have been
adapted from other fields.
2.2.1 Benchmark Methods & Systems
Systems that have been developed for syndromic
surveillance analysis include aberrancy-detection al-
gorithms. The Early Aberration Reporting System
(EARS) uses a variety of statistical aberration detec-
tion methods that have been developed by epidemi-
ologists to provide analysis for public health surveil-
lance data (Hutwagner et al., 2003). Another well-
known system for surveillance analysis is the Real-
time Outbreak and Disease Surveillance (RODS) sys-
tem. This system relies on real-time data collection
(Tsui et al., 2003). The data is saved to a database
where it then undergoes data warehousing techniques
to set up the data for analysis. The data is then an-
alyzed through various statistical aberrancy-detection
algorithms (Tsui et al., 2003). What’s strange about
recent events (WSARE) utilizes a bayesian network
to produce a baseline distribution that is then used to
compare against data (Wong et al., 2005). The soft-
ware SatScan analyzes spatial, temporal and space-
time scan statistics using the poisson or bernoulli
model based on requirements specified by the user
(Kulldorff, 2010).
2.2.2 Knowledge-based Systems
Syndromic surveillance requires the need for describ-
ing concepts, properties and relationships involved
in the process of data collection, analysis and re-
sult interpretation in order for a timely and accurate
evaluation to be performed (Buckeridge et al., 2002;
Buckeridge et al., 2008; Collier et al., 2010; Okhma-
tovskaia et al., 2009; Chapman et al., 2010). Ontolo-
gies are useful for describing, classifying and cate-
gorizing data. Due to this, a variety of ontologies and
ontology-based systems have been developed to aid in
the field of syndromic surveillance. Some of the sys-
tems currently using ontologies includes bioSTORM
and BioCaster (Buckeridge et al., 2002; Buckeridge
et al., 2008; Collier et al., 2010).
BioSTORM (Biological Spatio-Temporal Out-
break Reasoning Module) is a software system which
aims at providing a variety of analysis techniques and
rapidly integrating a diverse data set in order to pro-
cess data analysis in a timely manner (Buckeridge
AnOntology-basedFrameworkforSyndromicSurveillanceMethodSelection
397

et al., 2002; OConnor et al., 2003). It contains three
ontologies in its framework: the data-source ontology,
the problem-solving ontology and the data-mapping
ontology. As discussed in Section 2, incoming data
can range from a variety of different sources, it is
common practice to gather a large amount of data
from all these sources in order to make-up for the
loss of specificity within the data (Buckeridge et al.,
2008). The data-source ontology aims at describ-
ing and unifying data from various sources and data
streams (Buckeridge et al., 2002; OConnor et al.,
2003). The problem-solving ontology contains a li-
brary of statistical based and knowledge based prob-
lem solvers for analyzing data (Buckeridge et al.,
2008). The problem solving methods are categorized
and annotated in the ontology. Lastly, the mapping
ontology, aims at providing the correct problem solv-
ing technique to use for a set of data source which
will result in efficient data analysis(Buckeridge et al.,
2002; OConnor et al., 2003).
BioCaster is an ontology-driven system which
provides internet surveillance for potential outbreaks
found through global news reports (Collier et al.,
2010). The BioCaster ontology (BCO) aims at de-
scribing relations between terms in order to detect and
risk assess public health events, bridge the gap be-
tween (multilingual) grey literature and existing stan-
dards in biomedicine, mediate integration of content
across languages, and be available open source (Col-
lier et al., 2010).
Other ontologies that have been composed for
use in syndromic surveillance include: the syn-
dromic surveillance ontology (SSO) and the popu-
lation health ontology (Okhmatovskaia et al., 2009;
Chapman et al., 2010; Buckeridge et al., 2002). The
SSO aims at standardizing surveillance syndromes
and providing a classification of these syndromes
(Okhmatovskaia et al., 2009; Chapman et al., 2010).
The population health ontology describes how popu-
lation level health data relate to the underlying state
of illness in a population (Buckeridge et al., 2002).
This ontology describes determinants of disease, dis-
ease, illness as well as temporal and spatial changes
in determinants, disease and illness (Buckeridge et al.,
2002).
3 PROPOSAL & DISCUSSION
The system aims at providing a recommendation of
methods to be used for syndromic surveillance data
analysis in a descriptive manner to an end user. This
will thus allow a user to interpret the recommended
method without the need of a technical background.
The methods provided will be recommended statis-
tics, algorithms or systems which can be used to effi-
ciently detect a disease outbreak within a set of data.
The ontology will reason based on a set of parameters
provided by the user. Some of the parameters that
must be taken into consideration when developing an
ontology to describe an algorithm would be the data
source and input format, expected output format and
variables of importance to the end-user such as per-
formance, time, quality, and trust.
3.1 User Perspective
Systems are composed of many types of users rang-
ing from novice to experts. In the case of syndromic
surveillance, a typical end-user consist of a health an-
alyst or epidemiologist who analyzes a data set and
determines whether a disease outbreak is occurring.
There are various different methods which can be
used for conducting this analysis, some of these meth-
ods have been discussed in section 2.2. In order to de-
termine which method is best suited for a set of data,
the user performing the analysis would have to be an
expert in all systems. This is usually not the case,
for example, the user may be knowledgeable in var-
ious statistical methods which exist for analysis, but
may not consider other methods such as neural net-
works or genetic algorithms since they may not have
sufficient background in the area to understand these
algorithms.
Determining which analysis approach to take is
also dependent on requirements specified by the user.
These requirements can be defined based on what the
user believes will bring the most value to their analy-
sis. For example, a user can set one of their require-
ments as being performance measure. Different data
sets may require a different definition of performance;
OTC data can rely on how fast an outbreak was de-
tected, or the accuracy with which it was detected by
looking at false positives and false negatives attained
during the process. While ED data could also rely on
the timeliness of detection but also the speed at which
the geographical location of the outbreak was found
to occur.
It is important to consider a users perspective
when determining an algorithm best suited for anal-
ysis on a type of data-set. Defining the need of the
user will aid in bringing value to their analysis. This
need will be defined through requirements, presented
as input parameters in the proposed system.
3.2 Leveraging Knowledge
Leveraging knowledge describes how the transfer of
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
398
knowledge between two people is bi-directional and
that “knowledge grows when used and depreciates
when unused” (Firm et al., 2000). In order to take
full advantage of the existing syndromic surveillance
methods, the notion of leveraging knowledge is im-
portant to consider. For example, an epidemiologist
may look at data and determine a variety of bench-
mark statistics that they can pass through the data,
while a computer scientist could look at the same set
of data and come up with a list of algorithms which
could render interesting results. The epidemiologist
would not know to consider these algorithms before-
hand, and may not have a full-understanding of the
advantages they provide because they would not have
the technological background required.
For a system to be effective, it must be able to
eliminate the barrier formed and incorporate this no-
tion of bi-directional knowledge sharing. In other
words, by having the system describe each method
in a descriptive manner will aid in eliminating any in-
terpretation barrier previously encountered.
3.3 System Architecture
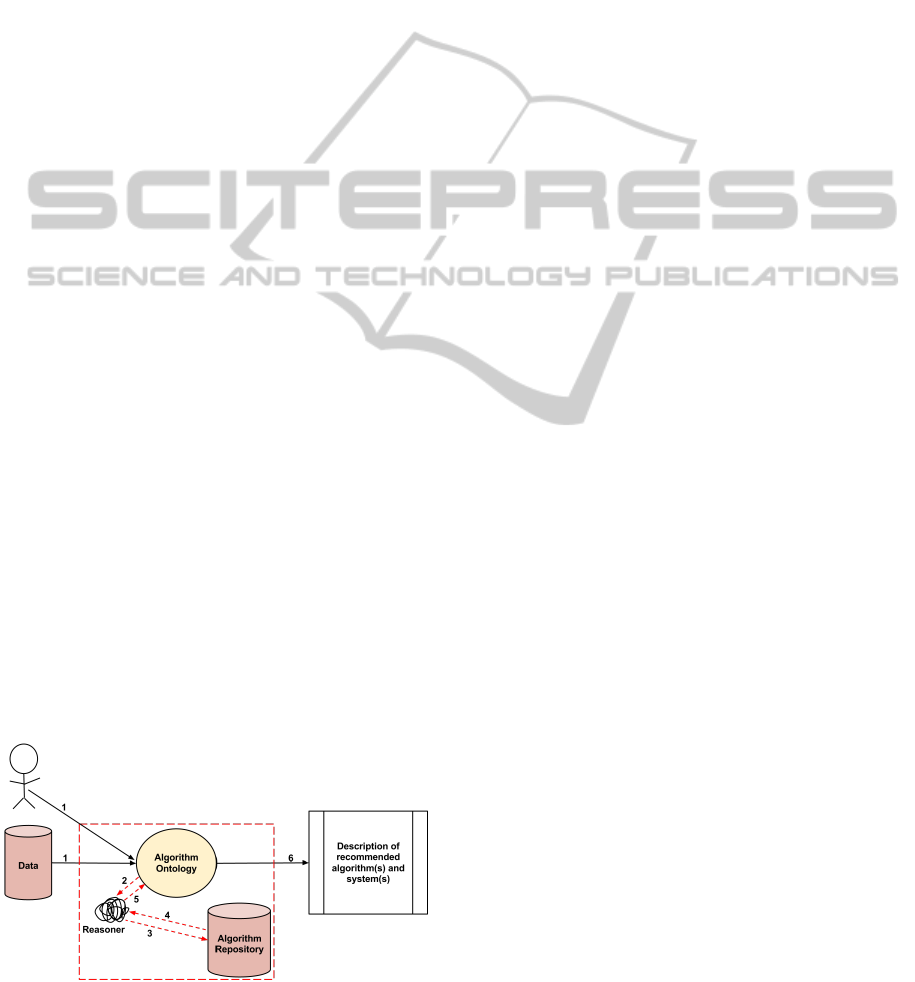
Figure 1 displays the proposed system architecture for
the procedure of gathering a set of methods best suited
for the data being analyzed. The following steps de-
scribe the overall process of the system.
1. Data is passed to the algorithm ontology. The data
includes information about the data specifying pa-
rameters such as input and expected output.
2. The reasoner classifies the data based on relation-
ships defined within the ontology.
3. A repository containing descriptions of algo-
rithms and systems is queried for the best suited
method(s) given the specifications provided.
4. & 5. A set of methods to use for analysis is at-
tained.
6. The recommended methods are described to the
user.
Figure 1: Proposed system architecture.
4 FUTURE WORK
The current proposed system evolves around an al-
gorithm ontology. This ontology will interpret a set
of parameters attained from an end-user, and recom-
mend method(s) best suited for the data set to be an-
alyzed. A better description of the parameters in-
volved is required for further development . As well,
a process for evaluating the system will also be in-
vestigated once further development has taken place.
Other factors will also be taken into consideration to
better the end-user experience, such as quality and
trust. Though the system will send a set of recom-
mended methods, the user would only use the method
if assured that it is reliable, and produces accurate re-
sults. Research will also be done on how to incorpo-
rate other existing ontologies to the system architec-
ture, such as the data source ontology found in BioS-
TORM, or the syndromic surveillance ontology.
ACKNOWLEDGEMENTS
I would like to thank Deb Stacey for her support and
guidance with this work.
REFERENCES
Buckeridge, D. L., Graham, J. K., O’Connor, M. J., Choy,
M. K., Tu, S., and Musen, M. A. (2002). Knowledge-
based bioterrorism surveillance. In AMIA Symp, pages
76–80.
Buckeridge, D. L., Okhmatovskaia, A., Tu, S., O’Connor,
M., Nyulas, C., and Musen, M. A. (2008). Un-
derstanding detection performance in public health
surveillance: Modelling aberrancy-detection algo-
rithms. Journal of the American Medical Informatics
Association, 15:760–769.
Chapman, W. W., Dowling, J. N., Baer, A., Buckeridge,
D. L., Cochrane, D., Conway, M. A., Elkin, P., Espino,
J., Gunn, J. E., Hales, C. M., Hutwagner, L., Keller,
M., Larson, C., Noe, R., Okhmatovskaia, A., Olson,
K., Paladini, M., Scholer, M., Sniegoski, C., Thomp-
son, D., and Lober, B. (2010). Developing syndrome
definitions based on consensus and current use. Jour-
nal of the American Medical Informatics Association,
17:595–601.
Collier, N., Goodwin, R. M., McCrae, J., Doan, S., Kawa-
zoe, A., Conway, M., Kawtrakul, A., Takeuchi, K.,
and Dien, D. (2010). An ontology-driven system for
detecting global health events. In Proceedings of the
23rd International Conference on Computational Lin-
guistics, COLING ’10, pages 215–222, Stroudsburg,
PA, USA. Association for Computational Linguistics.
Crubezy, M., O’Connor, M., Pincus, Z., Musen, M. A., and
Buckeridge, D. L. (2005). Ontology-centered syn-
AnOntology-basedFrameworkforSyndromicSurveillanceMethodSelection
399

dromic surveillance for bioterrorism. IEEE Intelligent
Systems, 20:26–35.
Firm, T., Chain, V., and Network, V. (2000). Ten Ways to
Leverage Knowledge for Creating Value. Knowledge
Creation Diffusion Utilization.
Guthrie, G., Stacey, D. A., Calvert, D., and Edge, V.
(2005). Detection of disease outbreaks in pharmaceu-
tical sales: Neural networks and threshold algorithms.
Public Health, pages 3138–3143 ST – Detection of
disease outbreaks in.
Henning, K. (2004). What is syndromic surveillance?
MMWR Morbidity and Mortality Weekly Report,
53:5–11.
Hutwagner, L., Thompson, W., Seeman, G. M., and Tread-
well, T. (2003). The bioterrorism preparedness and
response early aberration reporting system (EARS).
Journal of urban health bulletin of the New York
Academy of Medicine, 80:i89–i96.
Kulldorff, M. (2010). SatScan user guide.
Kulldorff, M., Heffernan, R., Hartman, J., Assuno, R.,
and Mostashari, F. (2005). A spacetime permutation
scan statistic for disease outbreak detection. PLoS
Medicine, 2(3):e59.
Lu, H.-M., Zeng, D., Trujillo, L., Komatsu, K., and Chen,
H. (2008). Ontology-enhanced automatic chief com-
plaint classification for syndromic surveillance. Jour-
nal of Biomedical Informatics, 41:340–356.
McDade, J. and Franz, D. (1998). Bioterrorism as a public
health threat. Emerging Infectious Diseases, 4:488–
492.
Okhmatovskaia, A., Chapman, W., Collier, N., Espino, J.,
and Buckeridge, D. L. (2009). SSO: The syndromic
surveillance ontology. In Proc International Society
for Disease Surveillance, page (in press).
OConnor, M. J., Buckeridge, D. L., Choy, M., Crubezy,
M., Pincus, Z., and Musen, M. A. (2003). BioS-
TORM: A system for automated surveillance of di-
verse data sources. AMIA Annual Symposium pro-
ceedings, 2003:1071.
Tsui, F.-C., Espino, J. U., Dato, V. M., Gesteland, P. H.,
Hutman, J., and Wagner, M. M. (2003). Technical de-
scription of RODS: a real-time public health surveil-
lance system. Journal of the American Medical Infor-
matics Association, 10:399–408.
Wong, W.-K., Moore, A., Cooper, G., and Wagner, M.
(2005). What’s strange about recent events (wsare):
An algorithm for the early detection of disease out-
breaks. J. Mach. Learn. Res., 6:1961–1998.
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
400
