
PUBMED DATASET: A JAVA LIBRARY FOR AUTOMATIC
CONSTRUCTION OF EVALUATION DATASETS
Kirill Lassounski
1
, Sahudy Montenegro Gonz
´
alez
2
, Annabell del Real Tamariz
1
and Gabriel Lima de Oliveira
1
1
State University of Norte Fluminense, Campos dos Goytacazes, Brazil
2
Federal University of S
˜
ao Carlos, Sorocaba, Brazil
Keywords:
Information Retrieval, Text Mining, PubMed, Evaluation Dataset, Java.
Abstract:
The NCBI (National Center for Biotechnology Information) provides information about genes, proteins, scien-
tific literature, molecular structures among other resources related to bio-medicine. The NCBI has a database
called PubMed that stores about 21 millions of scientific articles. There are many researches in the information
retrieval field that need to automatically obtain useful data from PubMed to perform evaluation and testing.
This work describes a Java library to construct datasets, so that numerous scientific researches could evaluate
their results easily and quickly. Users must set input and output parameters such as article’s attributes (title,
abstract, keywords, etc.) to conform the dataset constructed as a serializable file. The creation of PubMed
Dataset came from the fact that the authors needed to build their own datasets to evaluate their system results.
In this article it is also presented the BioSearch Refinement system as a case study. The system utilizes the
library to construct the datasets used to evaluate its algorithm for automatic extraction of keyphrases. We also
discuss the benefits obtained from the usage of the PubMed Dataset.
1 INTRODUCTION
The NCBI website has in its PubMed database
around 21 million scientific articles, protein se-
quences, genomes and other. The database receives
more than 70 million queries every month. In a search
using the term ”brain disease”, 129.977 complete ar-
ticles were retrieved, of which approximately 25%
were published in the last three years. These articles
are examined daily by researchers from all over the
world looking for information that will help them in
their studies. As a result, there are a large number of
research projects in information retrieval that help to
categorize, cluster and describe these articles, provid-
ing quality tools for researchers.
This paper aims to describe the PubMed Dataset,
a Java library for the automatic creation of scientific
datasets on PubMed database. It offers to bioinfor-
matic researchers a valuable tool for the creation of
datasets to test their proposals. Users must set input
and output parameters such as article’s attributes (ti-
tle, abstract, keywords, etc.) to conform the dataset
constructed as a Java serializable file. It is impor-
tant to highlight that this proposal came from the fact
that the authors needed to build their own datasets to
evaluate two researches associated with the NCBI and
PubMed. A relevant factor for its development was
the considerable time spent on manual or semiauto-
matic construction of datasets to evaluate the research
results.
To augment the relevance of the PubMed Dataset
library in the world of bioinformatics, one can cite
(Feldman and Sanger, 2007), who point out that much
researches and applications of text mining in this area
exploit the data centered on the NCBI website, as it
represents the largest online repository of scientific
papers in the area of biomedicine published in En-
glish and other languages. In a complementary way,
(Krallinger et al., 2008) reviewed the literature until
2008, describing more than fifty articles with focus
on efficient retrieval and text mining techniques on
biological databases.
In this work it is also presented a case study of
the PubMed Dataset library. The case study is based
on a Web system called BioSearch Refinement which
proposes an algorithm for automatic extraction and
summarization of keyphrases. The system utilizes the
library to construct the datasets used to evaluate the
keyphrase extraction algorithm.
This paper is organized as follows. Section 2
343
Lassounski K., Montenegro González S., del Real Tamariz A. and Lima de Oliveira G..
PUBMED DATASET: A JAVA LIBRARY FOR AUTOMATIC CONSTRUCTION OF EVALUATION DATASETS.
DOI: 10.5220/0003797203430346
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2012), pages 343-346
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

presents a short review of related work. Section 3 de-
scribes the PubMed Dataset library and its features. In
Section 4, it is introduced the BioSearch Refinement
system, the construction process of two datasets used
for evaluation purposes and the benefits obtained from
the usage of PubMed Dataset are discussed. Section
5 presents final considerations of the work.
2 RELATED WORK
(Krallinger et al., 2008) point out that developers of
text mining applications need to accomplish mean-
ingful system evaluations and comparative studies.
They also call attention to the difficulty in construct-
ing proper evaluation datasets.
Many works built their evaluation datasets man-
ual or semi-automatically, collecting documents from
a variety of areas, such as scientific literature, news
articles, magazine articles. Some efforts to collect
documents are described in (Nguyen and Kan, 2007),
(Wan and Xiao, 2008) and (Medelyan, 2009). The
first constructed a dataset of 250 PDF documents us-
ing the Google SOAP API to find them. These docu-
ments were manually restricted to articles published
in scientific conferences, later converted to plain
text. The second manually annotated the DUC2001
dataset, consisting of 309 news articles collected from
TREC-9. The annotation process lasted two weeks.
One of the research contributions in the latter was
an automatically extracted corpus of 180 science re-
search papers from collaboratively tagged data on the
bookmarking website CiteULike.org.
Systems described in (Zaremba et al., 2009),
(Yang et al., 2010) and (Uddin et al., 2010) need to
test results based on data extracted from PubMed and
are evidences of the importance of having a tool for
building datasets.
3 PUBMED DATASET
DESCRIPTION
Each article in PubMed has a PMID identifier and is
described by several attributes such as title, abstract,
publication date, among others in a semi-structured
XML format. There were created several sets of tags,
but it is noteworthy that not all articles have all the
tags. The PubMed’s XML files follow the standard
of the NLM Journal Archiving and Interchange DTD
available at the National Library of Medicine.
3.1 Design Principles
The design principles that governed the development
of the library are: (1) compatibility/portability which
means that the library can be used on any operating
system through any Java code to build the necessary
datasets; (2) flexibility to easily adapt and configure
the input and output parameters by the users; and (3)
to be open source to allow the reuse and improvement
of the code by the bioinformatics community mem-
bers.
3.2 Goal and Features
The tool aims to provide bioinformatics researchers
with a resource to create test sets from data available
on the PubMed database. The datasets can be used on
qualitative and quantitative assessment of their pro-
posals.
Figure 1 shows the overall process of the PubMed
Dataset library. The class DownloadConfiguration
stores the input and output settings of the user. Once
the parameters are specified, the tool is ready to con-
nect to PubMed via Esearch. The PMIDs are ob-
tained from the server and the XML documents of the
articles are retrieved and placed into a DOM (Doc-
ument Object Model) using EFetch. The data re-
quested by the user is extracted from this structure.
ArticleDownloader is the class responsible for re-
trieving articles from PubMed.
Figure 1: Overview of the PubMed Dataset process.
The required input parameters are:
• the search term used to retrieve a set of articles
from the PubMed database;
• the maximum number of items in the dataset;
• the list of article’s attributes to conform the
dataset;
• a boolean flag mandatoryAttributes to specify
when an article will be consider or not to conform
the dataset. If it is set to true, an article with a
missing value for any attribute in the list will be
discarded.
The dataset is built according to the parameters in
the initial configuration. The output file containing
the dataset can be unserialized at any moment and the
dataset will be promptly available.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
344
4 BIOSEARCH REFINEMENT
SYSTEM
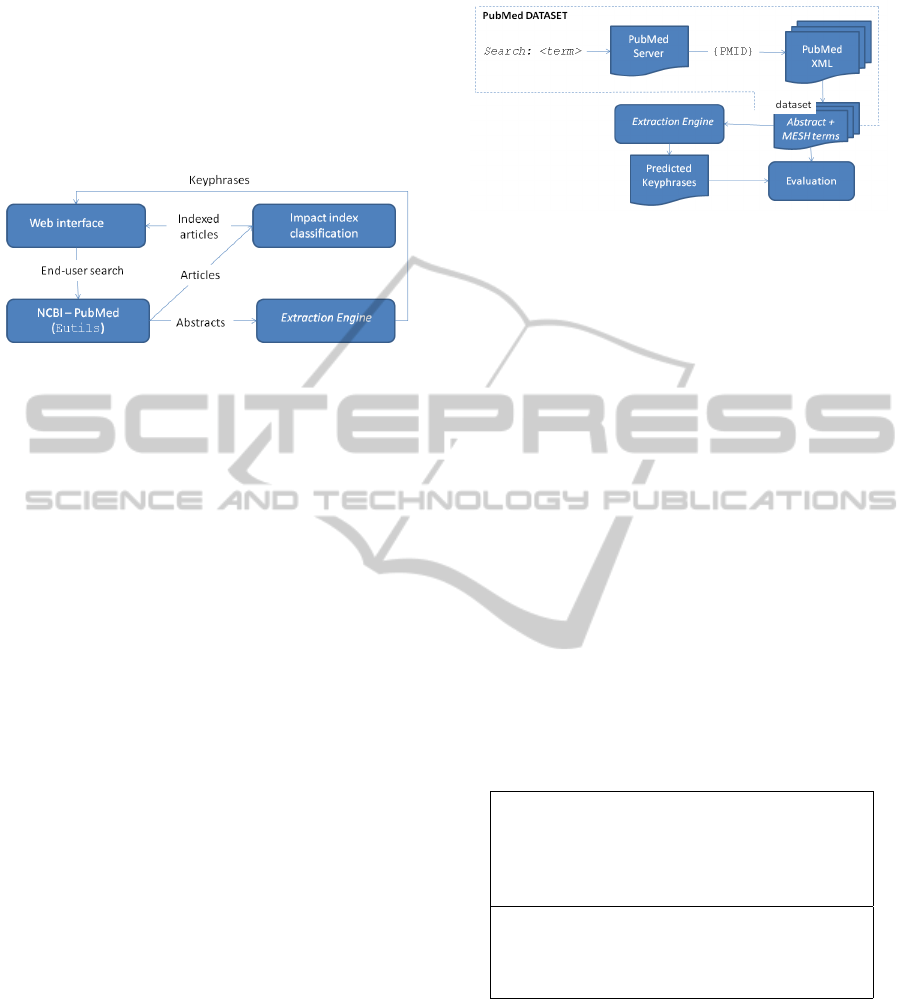
The BioSearch Refinement system is a Web applica-
tion developed in Java. Figure 2 shows the main func-
tionalities of the system.
Figure 2: BioSearch Refinement execution flowchart.
The keyphrases are extracted from the abstracts of
scientific papers returned from an initial query sub-
mitted on PubMed and are used to summarize the
main issues addressed by the retrieved papers. The
results can be filtered by keyphrases, helping the user
in their search for relevant material. The algorithm
to automatically obtain the keyphrases is called Ex-
traction Engine. It is based on concept matching and
refinement. To summarize the main subjects of the ar-
ticles, local keywords are obtained first and then dis-
tributed into concepts. After this, the most relevant
concepts from each article go to a global phase where
the final keyphrases are defined.
To validate the Extraction Engine proposal, it was
necessary to construct several datasets to evaluate
the quality of the extracted keyphrases in compar-
ison with manually tagged keyphrases. There are
two types of keywords on the PubMed database: the
MeSH index terms and the keywords provided by au-
thors. The experiments were conducted using the
MeSH terms because the author’s keywords are rarely
available in the database.
4.1 Experimental Evaluation
The experiment was accomplished to test the quality
of the datasets and measure execution times. To this
end, two datasets were created for the BioSearch Re-
finement system. The tests were performed on a 2.13
GHz Intel Core 2 Duo processor with 3GB of RAM.
First, the query terms were defined. Each dataset must
contain a set of abstracts that will be used to automati-
cally generate the keyphrases and the MeSH terms so
we can evaluate the quality of predicted keyphrases
using precision and recall metrics. The general pro-
cess of the experiment is shown in Figure 3.
Figure 3: Experiment dataset flowchart.
The code fragments below show a typical usage
of the library. The fragments specify the input/output
parameter configuration.
import static com.uenf.pubmeddataset.internet.ParameterName.*;
...
DownloadConfiguration config =
new DownloadConfiguration(ABSTRACT, PMID, MESH_TERMS);
ArticleDownloader downloader = new ArticleDownloader(config);
downloader.setMandatoryAttributes(true);
...
//building DATASET 1
List<DynaArticles> articles = downloader.getDynaArticles
("mycobacterium tuberculosis", 10000);
// the search term is treated as stopword
ConceptDataSet cds = new ConceptDataSet(articles,
"mycobacterium tuberculosis");
DataSetSerializer.serializeDataSet(cds, searchTerm);
...
// after unserialized, iterate over the dataset articles
Collection<DynaArticle> articles = cds.getArticles();
for(DynaArticle a:articles{
String ab = (String) a.getParameter(ABSTRACT).getValue();
System.out.println(ab);
}
...
//ConceptDataSet class extends DataSet abstract class and
// implements generateKeyWords abstract method
The results of the dataset construction are dis-
played on Table 1 below:
Table 1: Dataset configuration.
Dataset 1
search term: mycobacterium tuberculosis
max: 10.000 articles
Output file: data (abstract, MeSH terms)
from 8.045 PubMed articles
Dataset 2
search term: h1n1 influenza virus
max: 5.000 articles
Output file: data from 2.858 PubMed articles
Some reasons that caused the reduction of the
number of articles in the datasets were: (1) the
boolean flag was set to true; (2) articles without
MeSH terms were disregarded; (3) if the initial search
term appeared as a MeSH term, it was not included on
the dataset; and (4) if an article ran out of terms it was
also disregarded.
PUBMED DATASET: A JAVA LIBRARY FOR AUTOMATIC CONSTRUCTION OF EVALUATION DATASETS
345

4.2 Discussion about PubMed Dataset
Usage
Using the library for the creation of datasets was
quick and easy, simply by specifying the initial pa-
rameters. The construction of a dataset becomes very
flexible because its data and parameters can be eas-
ily modified and run again. The number of retrieved
articles will vary according to the attributes that were
specified by the user. The greater the number of at-
tributes the fewer items will be retrieved as the chance
of an article having all the attributes is smaller. The
quality of the dataset depends on what is available on
PubMed, because the datasets are an expression of the
data contained in the source. The performance results
are illustrated on Table 2.
Table 2: Performance times of PubMed Dataset.
Dataset Steps Runtime
1 Downloading 7,45 minutes
Serialization 2 seconds
Unserialization 3 seconds
Total 9,45 minutes
2 Downloading 3,11 minutes
Serialization 859 milliseconds
Unserialization 1 second
Total 3,13 minutes
The download phase is executed once and always
will be dependent on network traffic. In the case of
Dataset 1, the time for downloading 10.000 articles
was considerable high. Once the dataset is written to
disk, the time taken to load the dataset is very short,
making its use very convenient for testing.
5 CONCLUSIONS
By the team’s experience the time spent to build and
manage evaluation datasets is significant when com-
pared to the research itself, giving a positive bal-
ance to the use of this library. The PubMed Dataset
shows itself very useful on the fast, easy and effi-
cient construction of datasets. The three design prin-
ciples: portability, flexibility and to be open source
were achieved with success. Flexibility is a very im-
portant point that needs to be highlighted since any
article attribute can be included in the dataset.
It was presented a case study to create two datasets
using the library. The initial configuration was sim-
ply specified via Java code. The dataset is created
as a serialized file and the performance times of the
complete execution are not an issue. Yet, we believe
that future use of the library by other users will bring
new suggestions on how to improve the library to be
even more flexible and adaptable. The API’s source
code and documentation is available for download at
https://github.com/lassounski/PubMed-Dataset.
On an evaluation environment the tests are grad-
ually created and are often added more specific test
cases. Given this scenario, it is obvious the fact that
the tests will run several times. Without the use of this
library, the time spent obtaining test data and running
the tests would be remarkable. Note that the dataset
integrity and consistency are not guaranteed since the
data retrieved from PubMed may have missing values
and may vary over time.
ACKNOWLEDGEMENTS
This research was supported by CNPq (National
Counsel of Technological and Scientific Develop-
ment)/Brazil.
REFERENCES
Feldman, R. and Sanger, J. (2007). The Text Mining Hand-
book: Advanced Approaches in Analyzing Unstruc-
tured Data. Cambridge University Press.
Krallinger, M., Valencia, A., and Hirschman, L. (2008).
Linking genes to literature: text mining, informa-
tion extraction, and retrieval applications for biology.
Genome Biology, 9(2).
Medelyan, O. (2009). Human-competitive automatic topic
indexing. PhD thesis, University of Waikato.
Nguyen, T. and Kan, M. (2007). Keyphrase extraction
in scientific publications. In Proceedings of Interna-
tional Conference on Asian Digital Libraries (ICADL
’07), pages 317–326.
Uddin, J., Abulaish, M., and Dey, L. (2010). A concept-
driven biomedical knowledge extraction and visual-
ization framework for conceptualization of text cor-
pora. Journal of Biomedical Informatics, 43(6):1020–
1035.
Wan, Y. and Xiao, J. (2008). Collabrank: towards a collabo-
rative approach to single-document keyphrase extrac-
tion. In Proceedings of COLING.
Yang, Z., Lin, H., and Li, Y. (2010). Bioppisvmextractor:
A protein-protein interaction extractor for biomedical
literature using svm and rich feature sets. Journal of
Biomedical Informatics, 43:88–96.
Zaremba, S., Ramos-Santacruz, M., Hampton, T., Shetty,
P., Fedorko, J., Whitmore, J., Greene, J., Perna, N.,
Glasner, J., Plunkett, G., Shaker, M., and Pot, D.
(2009). Text-mining of pubmed abstracts by nat-
ural language processing to create a public knowl-
edge base on molecular mechanisms of bacterial en-
teropathogens. BMC Bioinformatics, 10(1).
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
346
