
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC
LIKELIHOOD COMPUTATIONS
Fernando Izquierdo-Carrasco
1
, Julien Gagneur
2
and Alexandros Stamatakis
1
1
The Exelixis Lab, Scientific Computing Group, Heidelberg Institute for Theoretical Studies
Schloss-Wolfsbrunnenweg 35, D-69118 Heidelberg, Germany
2
European Molecular Biology Laboratory, Meyerhofstr. 1, 69117 Heidelberg, Germany
Keywords:
Memory versus runtime trade-offs, Phylogenetic likelihood function, RAxML.
Abstract:
The revolution in wet-lab sequencing techniques that has given rise to a plethora of whole-genome or whole-
transcriptome sequencing projects, often targeting 50 up to 1000 species, poses new challenges for efficiently
computing the phylogenetic likelihood function both for phylogenetic inference and statistical post-analysis
purposes. The phylogenetic likelihood function as deployed in maximum likelihood and Bayesian inference
programs consumes the vast majority of computational resources, that is, memory and CPU time. Here, we
introduce and implement a novel, general, and versatile concept to trade additional computations for memory
consumption in the likelihood function which exhibits a surprisingly small impact on overall execution times.
When trading 50% of the required RAM for additional computations, the average execution time increase
because of additional computations amounts to only 15%. We demonstrate that, for a phylogeny with n
species only log(n) + 2 memory space is required for computing the likelihood. This is a promising result
given the exponential growth of molecular datasets.
1 INTRODUCTION
The rapid accumulation of molecular sequence data
that is driven by novel wet-lab techniques poses new
challenges regarding the design of programs for phy-
logenetic inference that rely on computing the Phylo-
genetic Likelihood Function (PLF) for phylogenetic
inference or statistical post-analysis. For instance, we
are currently involved in two large-scale sequencing
efforts aimed at obtaining, whole genome data for
approximately 150 species and whole transcriptome
data for approximately 1000 species
1
.
In all popular Maximum Likelihood (ML) and
Bayesian phylogenetic inference programs, the PLF
dominates both, the overall execution time as well
as the memory requirements by typically 85% -
95% (Stamatakis and Ott, 2008).
Based on our interactions with the RAxML user
community, we find that memory shortages are in-
creasingly becoming a problem and represent the
main limiting factor for large-scale phylogenetic anal-
yses, especially at the Genome level. At the same
time, the amount of available genomic data is growing
1
Because of non-disclosure agreements we can not pro-
vide further details about these projects at present.
at a faster pace than RAM (Random Access Mem-
ory) sizes and RAM access speeds. For instance, to
compute the likelihood on a simulated DNA align-
ment with 1,481 species and 20,000,000 sites (cor-
responding roughly to the 20,000 genes in the hu-
man genome) on a single tree topology under a simple
statistical model of nucleotide substitution within 48
hours, 1TB of memory and a total of 672 cores are
required.
We have already assessed some potential solutions
to this problem in previous work:
1. Single Precision. Deploying single precision
arithmetics that can reduce memory requirements
by 50% but can potentially induce numerical in-
stability, especially on datasets with more than
1000 taxa (Berger and Stamatakis, 2010).
2. Gappy Alignments. Novel algorithmic solutions
which rely on dataset-specific properties, that is,
their efficiency/applicability depends on the spe-
cific properties of the Multiple Sequence Align-
ment (MSA) that is used as input (Stamatakis and
Alachiotis, 2010). In particular, these algorithmic
approaches do not work on multi-gene datasets
that do not contain enough missing data on a per-
gene basis, that is, when gene sequences for a
86
Izquierdo-Carrasco F., Gagneur J. and Stamatakis A..
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS.
DOI: 10.5220/0003765600860095
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2012), pages 86-95
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

certain number of taxa under study are not avail-
able. In other words, these approaches are not ap-
plicable to emerging, dense whole-genome MSAs
without missing data.
3. Out-Of-Core. We recently introduced a gen-
erally applicable method for saving memory,
that does not rely on MSA-specific characteris-
tics. We evaluated an out-of-core (frequently
also called external memory algorithm) approach
that reduces RAM requirements by moving data
back and forth between the RAM and the
disk (Izquierdo-Carrasco and Stamatakis, 2011).
However, the increase of at least one order of
magnitude in overall execution times because of
slow read/write operations to/from disk does not
yield this approach practical, even when a high
performance parallel file system is used.
Here, we introduce a novel and significantly more
efficient method that is based on trading computations
(runningtime) for memory. The method is completely
independent from the MSA structure and is therefore
particularly useful for reducing RAM requirements
of “dense” whole-genome alignments. Moreover, it
can be combined with the single precision and gappy
alignment memory saving approaches (see above) to
yield even further savings. When reducing the amount
of available RAM to 50% of the total RAM required
for computing the likelihood on a tree for a given
MSA, we obtain a respective slowdown (because of
additional (re-)computations) of only 15%. This rep-
resents a relatively small run-time increase that most
end-users will not even notice. On the other hand,
users will notice when their analyses crash because
they have run out of RAM or the OS (operating sys-
tem) starts paging.
The remainder of this paper is organized as fol-
lows: We briefly review related work in Section 2.
In Section 3 we describe the underlying idea for
the memory saving approach, demonstrate that only
log(n) + 2 vectors are required, and introduce two
vector replacement strategies. In Section 4 we de-
scribe the experimentalsetup and providecorrespond-
ing results. We conclude and discuss directions of fu-
ture work in Section 5.
2 RELATED WORK
One common approach for reducing memory require-
ments are out-of-core algorithms. These are specif-
ically designed to minimize the I/O overhead via
explicit, application-specific, data placement control
and movement. Out-of-core algorithms mostly aim at
optimizing data movement between disk and RAM.
A comprehensive review of work on out-of-core al-
gorithms can be found in (Vitter, 2008). In phylo-
genetics, out-of-core algorithms have been applied to
Neighbor-Joining (Wheeler, 2009; Martin Simonsen
and Pedersen, 2010). Moreover, we recently intro-
duced a generic out-of-core implementation for PLF
computations (Izquierdo-Carrasco and Stamatakis,
2011). As already mentioned, the performance of our
out-of-core implementation was rather disappoint-
ing and is therefore not applicable to likelihood-
based inference on dense whole-genome alignments.
Nonetheless, our approach was significantly more ef-
ficient than the paging strategy of the OS.
The term time-memory trade-off engineering
refers to situations/approaches where memory re-
quirements/utilization is reduced at the cost of ad-
ditional (re-)computations, that is, at the expense of
increased run time. This trade-off engineering ap-
proach has been applied in diverse fields such as lan-
guage recognition (Dri and Galil, 1984), cryptogra-
phy (Barkan et al., 2006), and packet scheduling (Xu
and Lipton, 2005).
We are not aware of any applications of or exper-
iments with time-memory trade-off engineering ap-
proaches in the field of computational phylogenetics.
3 RECOMPUTATION OF
ANCESTRAL PROBABILITY
VECTORS
3.1 PLF Memory Requirements
A detailed discussion on PLF Memory requirements
is provided in (Izquierdo-Carrasco and Stamatakis,
2011). We include a part of this discussion, that is
also relevant for the work presented here. The PLF
is defined on unrooted binary trees. The n extant
species/organisms of the MSA under study are lo-
cated at the tips of the tree, whereas the n − 2 in-
ner nodes represent extinct common ancestors. The
molecular sequence data in the MSA that has a length
of s sites (alignment columns) is located at the tips of
the tree. The memory requirements for storing those
n tip vectors of length s is not problematic, because
one 32-bit integer is sufficient to store, for instance, 8
nucleotides when ambiguous DNA character encod-
ing (requiring 4 bits to store one nucleotide) is used.
Hence, for the aforementioned large dataset (1,481
species, 20,000,000 sites) only 13GB are required to
store the actual sequence data compared to 1 TB for
storing ancestral probability vectors.
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS
87

Hence, the memory requirements of the PLF are
dominated by the space for storing the ancestral prob-
ability vectors that are located at the ancestral (inner)
nodes of the tree. Depending on the PLF implemen-
tation, at least one such vector (a total of n− 2) will
need to be stored per ancestral node. For each align-
ment site i,i = 1...s, an ancestral probability vector
needs to hold the data for the probability of observing
an
A, C, G
or
T
. Thus, under double precision arith-
metics and for DNA data a total of (n− 2)·8·4·s bytes
is required for the most simple statistical model of nu-
cleotide substitution. When the less simplistic, stan-
dard Γ model of rate heterogeneity (Yang, 1994) with
4 discrete rates is deployed, this number increases by
a factor of four ((n− 2) · 8 · 16 · s), since we need to
store 16 probabilities for each alignment site i. Fur-
thermore, if protein data is used (that has 20 instead of
4 states) in conjunction with the Γ model of rate het-
erogeneity, the memory requirements for storing an-
cestral probability vectors increase to (n− 2)· 8· 80· s
bytes.
To obtain the likelihood of a given, fixed tree, the
PLF performs a post-order tree traversal that starts at
a virtual root. Such a virtual root can be placed ar-
bitrarily into any branch of the unrooted tree without
changing the overall likelihood of the tree. By means
of the post-order tree traversal, the ancestral probabil-
ity vectors are computed bottom-up from the leaves
toward the virtual root.
For a more detailed description of PLF computa-
tions and efficient PLF implementations please refer
to (Stamatakis, 2011). For understanding the memory
access pattern during the post-order traversal, let us
consider equation (Felsenstein, 1981) of the Felsen-
stein pruning algorithm.
This equation computes the ancestral probability
vector entry
~
L
(p)
A
for observing the nucleotide
A
at site
i of a parent node p, with two child nodes l and r given
the respective branch lengths b
l
and b
r
, the corre-
sponding transition probability matrices P(b
l
), P(b
r
),
and the probability vectors of the children
~
L
(l)
,
~
L
(r)
:
~
L
(p)
A
(i) =
T
∑
S=A
P
AS
(b
l
)
~
L
(l)
S
(i)
T
∑
S=A
P
AS
(b
r
)
~
L
(r)
S
(i)
(1)
In order to compute L
(p)
A
(i), we do not necessarily
need to immediately have available in memory vec-
tors L
(l)
S
(i) and L
(r)
S
(i), since they can be obtained by a
recursive descent (a post-order subtree traversal) into
the subtrees rooted at l and r using the above equation.
In other words, if we do not have enough memory
available, we can recursively re-compute the values
of L
(l)
and L
(r)
. Note that, the recursion terminates
when we reach the tips (leaves) of the tree and that
memory requirements for storing tip vectors are neg-
ligible compared to ancestral vectors (see above).
If not all ancestral probability vectors fit in RAM,
the required vectors for the operation at hand can still
be obtained by conducting some additional compu-
tations for obtaining them by applying equation 1.
We observe that, when both L
(l)
S
(i) and L
(r)
S
(i) have
been used (consumed) for calculating L
(p)
A
(i), the two
child vectors are not required any more. That is, those
two vectors can be omitted/dropped from RAM or be
overwritten in RAM to save space. Therefore, the
likelihood of a tree can be computed without storing
all n − 2 ancestral vectors. Instead, a smaller amount
of space for only storing x < (n − 2) vectors can be
used. Those x vectors can then dynamically be as-
signed to a subset (varying over time) of the n − 2
inner nodes. This gives rise to the following question:
Given a MSA with n taxa, what is the minimum num-
ber of inner vectors x
min
that must reside in memory
to be able to compute the likelihood on any unrooted
binary tree topology with n taxa?
Due to the post-order traversal of the binary tree
topology, some ancestral vectors need to be stored
as intermediate results. Consider an ancestral node
p that has two subtrees rooted at child nodes l and r
with identical subtree depth. When the computations
on the first subtree for obtaining L
(l)
have been com-
pleted, this vector needs to be kept in memory until
the ancestral probability vector L
(r)
has been calcu-
lated to be able to compute L
(p)
.
1
2
3 4
5
6
7
virtual root
Figure 1: A balanced subtree, where the vector of inner
node 1 is oriented in the direction of the virtual root. In
order to compute the likelihood of vector 1, the bold vec-
tors must be held in memory. The transparent vectors may
reside in memory but are not required to.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
88

In the following section 3.2, we demonstrate that
the minimum number of ancestral vectors x
min
(n) that
must reside in RAM to be able to compute the likeli-
hood of a tree with n taxa is log
2
(n) + 2. For obtain-
ing this lower memory bound, we consider the worst
case tree shape, that is, a perfectly balanced unrooted
binary tree with the virtual root placed into the inner-
most branch.
Figure 1, where we first descended into the left
subtree, depicts this worst case scenario for n = 4.
The probability vectors will be written in the follow-
ing order:
3, 4, 2, 6, 7, 5, 1
. Figure 1 shows
the number of vectors required in memory (log
2
(4) +
2 = 4) at the point of time where vector 5 needs to be
computed. At any other point of time during the post-
order traversal, holding 3 vectors in RAM is sufficient
to successfully proceed with the computations.
3.2 Minimum Memory Requirements
The underlying idea of our approach is to reduce the
total number of ancestral probability vectors residing
in RAM at any given point of time. Let PLF be the
likelihood function (see Equation 1), which —for the
sake of simplifying our notation— can be computed
at a tip (essentially at zero computational cost) or an
inner node given the ancestral probability vectors of
its two child nodes. The PLF always returns an an-
cestral probability vector as result.
As a pre-processing step, we compute the num-
ber of descendants (size of the respective subtree) for
each inner node. This can be implemented via a sin-
gle post-order tree traversal. The memory required
for storing the number of descendants as integers at
each node is negligible compared to required proba-
bility vector space. Given this information, the binary
tree data structure can be re-ordered such that, for ev-
ery node p, the left child node l contains the larger
number of descendants and the right child node r con-
tains the smaller number of descendants. We can now
compute the PLF of the tree by invoking the follow-
ing recursive procedure f at the virtual root p of the
tree:
proc f(p) ≡
if isALeaf(p) then return(PLF(p)) fi;
v
l
:= f(p.l); Step 1
v
r
:= f(p.r); Step 2
return(PLF(v
l
,v
r
)) Step 3
.
During this computation, the maximum number of
probabilityvectors x
min
(n) that need to reside in mem-
ory for any tree with n leaves is log
2
(n) + 2. This up-
per bound is required if and only if the binary tree is
balanced.
Proof. We demonstrate the above theorem by re-
cursion. x
min
(1) = 1, since for a tree with a sin-
gle node, only the sequence data (a single proba-
bility vector) need to be stored. Now assume that
x
min
(n− 1) = log
2
(n− 1) + 2 and consider a tree with
n leaves. We execute all steps of f(p), where p is
the root, and keep track of the number of probability
vectors that are stored simultaneously in RAM:
• Step 1. Computing f(p.l) requires storing at
most x
min
(n − 1) probability vectors simultane-
ously which is strictly less than log
2
(n)+ 2. Once
f(p.l) has been computed only the result vector is
stored (i.e., only one single probability vector).
• Step 2. Because the subtree size of the right de-
scendant of the root p.r must be ≤ n/2, this com-
putation needs to simultaneously store at most
log
2
(n/2) + 2 = log
2
(n) + 1 probability vectors.
Here, we need to add the number of probability
vectors that required to be maintained in RAM af-
ter completion of Step 1. Thus, overall, Step 2
needs to simultaneously hold at most log
2
(n) + 2
probability vectors in RAM. Once the results of
f(p.l) and f(p.r) have been computed we are left
with two probability vectors v
l
,v
r
that need to re-
side in memory.
• Step 3. This step requires 3 probability vectors to
be stored at the same time, namely, v
l
, v
r
, and the
vector to store the result of PLF(v
l
,v
r
). Note that,
to obtain the overall likelihood of the tree (a single
numerical value), we need to apply a function g()
to the single result vector returned by f(), that is,
g( f(p)). Function g() simply uses the data in the
result vector to calculate the likelihood score over
all root vector entries.
Hence the peak in memory usage is reached dur-
ing Step 2 and its upper bound is log
2
(n) + 2. More-
over, this upper bound is reached if and only if the
number of descendants in the respective left and right
subtrees is identical for all nodes, that is, for a bal-
anced tree.
3.3 Basic Implementation
While holding log
2
(n) + 2 vectors in RAM provides
sufficient space for computing the PLF, this will in-
duce a significant run-time increase (due to recom-
putations) compared to holding n vectors in memory.
In practice, we need to analyze and determine a rea-
sonable run-time versus RAM trade-off as well as an
appropriate vector replacement/overwriting strategy.
For instance, in the RAxML or MrBayes PLF im-
plementations, some ancestral vectors (depending on
their location in the tree) can be recomputed faster
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS
89

than others. In particular, cases where the left and/or
right child vectors are tip sequences can be handled
more efficiently. For instance, an observed nucleotide
A
at a tip sequence corresponds to a simple probability
vector of the form [P(A) := 1.0, P(C) := 0.0,P(G) :=
0.0,P(T) := 0.0]. This property of tip vectors can be
used for saving computations in equation 1.
Also, typical topological search operators for find-
ing/constructing a tree topology with an improved
likelihood score such as SPR (Subtree Pruning and
Re-grafting), NNI (Nearest Neighbor Interchange) or
TBR (Tree Bisection and Reconnection) only apply
local changes to the tree topology. In other words,
the majority of the ancestral probability vectors is not
affected by the topological change and does there-
fore not need to be recomputed/updated with respect
to the locally altered tree topology via a full post-
order tree traversal. Therefore, if all ancestral vec-
tors reside in RAM, only a very small part of the
tree needs to be (re-)traversed after a SPR move, for
instance. All standard ML-based programs such as
GARLI (Zwickl, 2006), PHYML 3.0 (Guindon et al.,
2010), and RAxML (Stamatakis, 2006) deploy search
strategies that typically require updating only a small
fraction of probability vectors in the vicinity of the
topological change.
Therefore, devising an appropriate strategy (see
Section 3.4) for deciding which vectors shall remain
in RAM and which can be discarded (because they
can be recomputed at a lower computational cost)
can have a substantial impact on the induced run
time overhead when holding, for instance, x := n/2
vectors in RAM. In the following, we will outline
how to compute the PLF and conduct SPR-based tree
searches with x < n vectors in RAM.
Let n be the number of species, n− 2 the number
of ancestral probability vectors, and x the number of
available slots in memory, where log
2
(n) + 2 ≤ x <
n (i.e., n − x vectors are not stored, but recomputed
on demand). Let w be the number of bytes required
for storing an ancestral probability vector (all vectors
have the same size). Our implementation will only
allocate x· w bytes, rather than n· w. We henceforth
use the term slot to denote a RAM segment of w bytes
that can hold an ancestral probability vector.
We define the following C data structure (details
omitted) to keep track of the vector-to-slot mapping
of all ancestral vectors and for implementing replace-
ment strategies:
typedef struct
{
int numVectors;
size_t width;
double **tmpvectors;
int *iVector;
int *iNode;
int *unpinnable;
boolean allSlotsBusy;
unpin_strategy strategy;
}recompVectors;
The array
tmpvectors
is a list of pointers to
a set of slots (i.e., starting addresses of allocated
RAM memory) of size
numVectors
(x) and width
numVector
(w). The array
iVector
has length x and
is indexed by the slot id. Each entry holds the node
id of the ancestral vector that is currently stored in the
indexed slot. If the slot is free, the value is set to a
dedicated
SLOT_UNUSED
code. The array
iNode
has
length n− 2 and is indexed using the unique node ids
of all ancestral vectors in the tree. When the corre-
sponding vector resides in RAM, its array entry holds
the corresponding slot id. If the vector does not re-
side in RAM the array entry is set to the special code
NODE_UNPINNED
. Henceforth, we denote the avail-
ability/unavailability of an ancestral vector in RAM
as
pinned/unpinned
The array
unpinnable
tracks
which slots are available for unpinning, that is, which
slots that currently hold an ancestral vector can be
overwritten, if required.
The set of ancestral vectors that are stored in the
memory slots changes dynamically during the com-
putation of the PLF (i.e., during full tree traversals
and tree searches). The pattern of dynamic change in
the slot vector also depends on the selected recom-
putation/replacement strategy. For each PLF invoca-
tion, be it for evaluating a SPR move or completely
re-traversing the tree, the above data structure is up-
dated accordingly to ensure consistency.
Whenever we need to compute a local tree traver-
sal (following the application of an SPR move) to
compute the likelihood of the altered tree topology,
we initially just compute the traversal order which is
part of the standard RAxML implementation. The
traversal order is essentially a list that stores in which
order ancestral probability vectors need to be com-
puted. In other words, the traversal descriptor de-
scribes the partial or full post-order tree traversal re-
quired to correctly compute the likelihood of a tree.
For using x < n vectors, we introduce a so-called
traversal order check, which extends the traversal
steps (the traversal list) that assume that all n vec-
tors reside in RAM. By this traversal order extension,
we guarantee that all missing vectors (not residing in
RAM) will be recomputed as needed. The effect of
reducing the number of vectors residing in RAM is
that, traversal lists become longer, that is, more nodes
are visited and thereby run time increases. When the
traversal is initiated, all vectors in the traversal list
that already reside in RAM (they are pinned to a slot)
are protected (marked as
unpinnable
) such that, they
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
90

will not be overwritten by intermediate vectors of the
recomputation steps.
If an ancestral vector slot needs to be writ-
ten/stored by the traversal, there are three cases:
1. The vector resides in a slot (already in memory).
We can just read and/or write to this slot.
2. The vector is not pinned, but there exists a free
slot, which is then pinned to this vector.
3. The vector is not pinned and there is no free slot
available. A residing vector must be replaced and
the corresponding slot needs to be pinned to the
required vector.
Since the traversal only visits each vector at most
once, the corresponding children of this vector can be
unpinned once it has been written to memory. Instead
of unpinning them directly, they are merely marked
for unpinning. The real overwrite only takes place
if the slot is selected by the replacement strategy for
storing another vector. Otherwise, the slot will store
the values of the current vector for as long as possible
for potential future re-use.
3.4 Replacement Strategies
In analogy to the replacement strategies discussed in
the out-of-core implementation (Izquierdo-Carrasco
and Stamatakis, 2011), there are numerous ap-
proaches for deciding which memory slot should be
overwritten by a new ancestral vector that does cur-
rently not reside in RAM. We implement and analyze
the following two replacement strategies.
Random. A random slot not flagged as pinned is se-
lected. The random strategy is a na¨ıve approach
with minimal overhead and is used as baseline for
performance comparisons.
MRC. Minimum Recomputation Cost. The slot with
minimum subtree size (see below) and not flagged
as pinned is selected.
The MRC strategy entails a slight overhead for
keeping track of which vectors will be most expen-
sive to recompute and should therefore be kept in
RAM for as long as possible. Consider an unrooted
binary tree T with n tips. Each inner node i in an
unrooted binary tree with n taxa can be regarded as
a trifurcation that defines three subtrees T
i,a
, T
i,b
, and
T
i,c
corresponding to the three outgoing branches a,b,
and c. Given a subtree T
i,x
, we define its subtree
size sts(T
i,x
) as the number of tips beneath inner node
i in the direction of the outgoing branch x. Thus,
sts(T
i,a
) + sts(T
i,b
) + sts(T
i,c
) = n holds for any in-
ner node i in an unrooted binary tree with n taxa/tips.
When conducting likelihood computations, the tree is
always rooted by a virtual root. Hence, if the virtual
root is located in the direction of branch c the rele-
vant subtree size with respect to recomputation cost
at inner node i is sts(T
rooted
i
) := sts(T
i,a
) + sts(T
i,b
).
We use this rooted subtree size sts(T
rooted
i
) to deter-
mine the recomputation cost for each ancestral vector
i given a virtual rooting of the tree. In particular, the
case sts(T
rooted
i
) = 2 (both children are tips) is par-
ticularly cheap to recompute, since tip vectors always
reside in RAM and recomputing ancestral vector i is
cheap (see above). In a perfectly balanced tree with
the root placed in the innermost branch, half of the
inner vectors have subtree size sts(T
rooted
i
) = 2.
As already mentioned, during a partial or full tree
traversal for computing the likelihood, all inner nodes
(vectors) involved are oriented in a given direction to-
ward the virtual root. Evidently, the subtree sizes will
change when the topology is altered and will need to
be updated accordingly.
In order to account for this, we keep an array of
subtree sizes, that is, for each inner node we store a
subtree size value. Whenever the topology changes, a
local traversal descriptor is created. This local traver-
sal descriptor starts at the new virtual root and recur-
sively includes the inner nodes whose orientation has
changed after the given topology change. Since this
exactly corresponds to the set of nodes whose subtree
size values must be updated, the subtree size array can
be updated via the same recursive descent.
3.5 Implementation Details
In the following we discuss some important details of
the recomputation process.
Largest Subtree First. The standard implementa-
tion of the PLF, where all ancestral vectors are avail-
able in memory, computes the PLF by conducting a
post-order traversal from an arbitrary rooting of the
tree. Thus, an ancestral probability vector can be
computed once the respective left and right child vec-
tors have been computed. In the standard RAxML
implementation, the traversal always recursively de-
scends into the left subtree first. The (arbitrary)
choice whether to descend into the left or right sub-
tree first, does not affect performance (nor the results)
when all ancestral vectors reside in RAM.
However, when not all vectors reside in RAM, the
choice whether to descend into the left or right subtree
first does matter. This is particularly important if we
use the minimum setting x := log
2
(n)+ 2, since other-
wise we may encounter situations where not enough
slots are available (see Section 3.2).
Suppose that, as in the standard implementation,
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS
91
1
2
5
4
virtual root
3
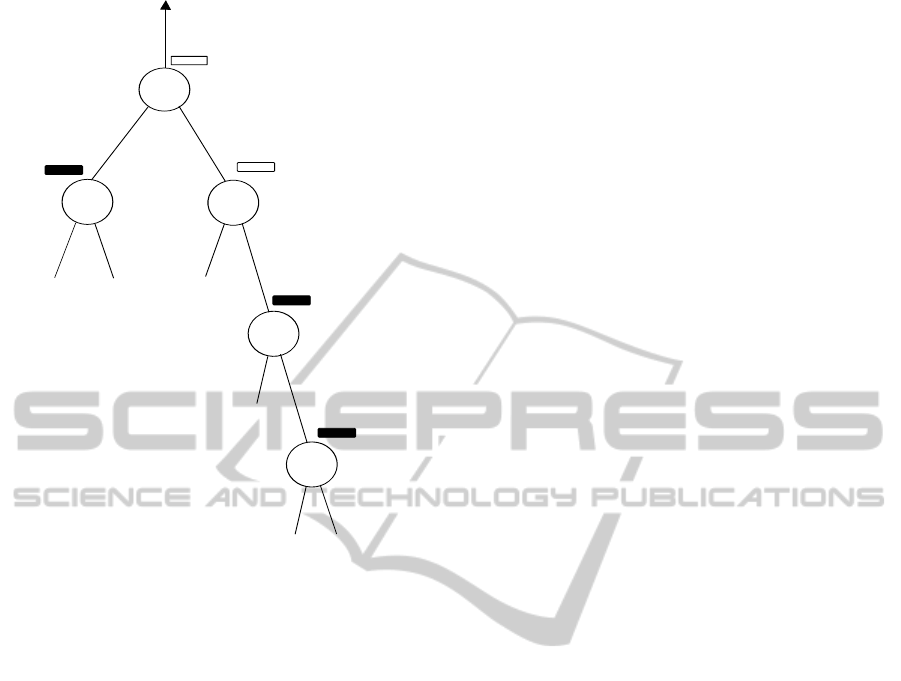
Figure 2: An unbalanced subtree, where the vector of inner
node 1 is oriented in the direction of the virtual root. Bold
rectangles represent vectors that must be held in memory if
we first descend into the left subtree and node 4 is being
written.
we always descend into the left subtree first. In the
example shown in Figure 2, the left subtree is signif-
icantly smaller than the right subtree. We would first
descend into the left subtree, which consists of a sin-
gle inner node. The child ancestral vector correspond-
ing to node 2 must be pinned to its slot. Thereafter,
we descend into the right subtree writing and pinning
nodes 3, 4, 5 (always assuming that we descend into
the left —smaller— subtree of eachnode first). While
we keep descending into the right subtree, the ances-
tral vector corresponding to node 2 remains pinned,
because it represents an intermediate result.
In contrast, if we initially descend into the right
subtree (which is always larger in the example), there
is no need to store intermediate results of the left sub-
tree (node 2). Also, nodes 4 and 5 can be imme-
diately unpinned as soon as their parent nodes have
been computed. Thus, by inverting the descent order
such as to always descend into the larger subtree first
(as required by our proof), we minimize the amount
of time for which intermediate vectors must remain
pinned. Note that, when two subtrees have the same
size, it does not matter into which subtree we descend
first.
If we descend into the smaller subtrees first, there
will be more vectors that need to remain pinned for
a longer time. This would also reduce the effective
size of the set of inexpensive-to-recompute vectors
that shall preferentially be overwritten, because more
vectors that are cheap to recompute need to remain
pinned since they are holding intermediate results.
In this scenario more expensive-to-recompute vectors
will need to be overwritten in memory and dropped
from RAM.
Determining the appropriate descent order (largest
subtree first) is trivial and induces a low compu-
tational overhead. When the traversal list is com-
puted, we simply need to compare the subtree sizes
of child nodes and make sure to always descend into
the largest subtree first.
Priority List. For this additional optimization, we
exploit a property of the SPR move technique. When
a candidate subtree is pruned (removed from the cur-
rently best tree), it will be re-inserted into several
branches of the tree from which it was removed to
evaluate the likelihood of different placements of the
candidate subtree within this tree. In the course of
those insertions, the subtree itself will not be changed
and only the ancestral vector at the root of the subtree
will need to be accessed for computations. Hence, we
maintain a dedicated list of pruned candidate subtree
nodes (a unpinning priority list) that can be preferen-
tially unpinned. Because of the design of lazy SPR
moves in RAxML (similar SPR flavors are used in
GARLI and PHYML) those nodes (corresponding to
ancestral vectors) will not be accessed while the can-
didate subtree is inserted into different branches of the
tree. Once this priority list is exhausted, the standard
MRC recomputation strategy is applied.
Full Traversals. Full post-order traversals of the
tree are required during certain phases of typical phy-
logenetic inference programs, for instance when op-
timizing global maximum likelihood model param-
eters (e.g., the α shape parameter of the Γ distribu-
tion or the rates in a GTR matrix) on the entire tree.
Full tree traversals are also important for the post-
analysis of fixed tree topologies, for instance, to es-
timate species divergence times. Full tree traversals
represent a particular case because every inner vector
of the tree needs to be visited and computed. Hence,
the number of vectors that need to be computed under
our memory reduction approach is exactly identical to
the number of vectors that need to be computed under
the standard implementation. Thus, there is no need
for additional computations while a large amount of
memory can be saved. While full tree traversals do
not dominate run times in standard tree search algo-
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
92

rithms, they can dominate execution times in down-
stream analysis tools.
4 EXPERIMENTAL SETUP AND
RESULTS
We have implemented the techniques described in
Section 3 in RAxML-Light v1.0.4. RAxML-Light
is a strapped-down dedicated version of RAxML
intended for large-scale phylogenetic inferences
on supercomputers. It implements a light-weight
software-based checkpointing mechanism and offers
fine-grained PThreads and MPI parallelizations
of the PLF. It has been used to compute a tree
on a dense simulated MSA with 1481 taxa and
20,000,000 sites that required 1TB of RAM and
ran in parallel with MPI on 672 cores. While not
yet published, RAxML-Light is available as open-
source code at www.exelixis-lab.org/software.html
and at https://github.com/stamatak. The source
code of the memory saving technique presented
here is available for download at www.exelixis-
lab.org/software/spare.zip and is currently being
integrated into the RAxML-Light development
version (development snapshots available at
https://github.com/fizquierdo/vectorRecomp).
4.1 Evaluation of Recomputation
Strategies
The recomputation algorithm yields exactly the same
log likelihood scores for the PLF as the standard algo-
rithm. Thus, for validating the correctness of our im-
plementation, it is sufficient to verify that the result-
ing trees and log likelihood scores of a ML tree search
with the standard and recomputation implementations
are identical. The increase of total run time depends
on the number x of inner vectors that are held in
memory and on the chosen unpinning strategy (MRC
versus RANDOM). We used indelible (Fletcher and
Yang, 2009) to generate simulated MSAs of 1500,
3000, and 5000 species. All experiments described in
this section were conducted for these three datasets.
We only show representative results for one dataset
(1500 taxa, 575 base pairs).
The results for the other two simulated datasets
(3000 taxa with 774 base pairs and 5000 taxa with
1074 base pairs), as well as for a biological dataset
with 500 taxa, are analogous and can be found in
the supplementary on-line material at www.exelixis-
lab.org/publications.html.
We chose to use simulated datasets to assess per-
formance for convenience. Detailed information on
indelible parameters used for dataset generation are
included in the supplementary on-line material. For
our purely computational work it does not matter
whether simulated or real data is used.
Initially, we ran the Parsimonator program (avail-
able at www.exelixis-lab.org/software.html to gener-
ate 10 distinct randomized stepwise addition order
parsimony starting trees for each MSA. For each
starting tree, we then executed a standard ML tree
search with RAxML-Light 1.0.4 (sequential version
with SSE3 intrinsics) and ML tree searches with the
recomputation version for the two replacement strate-
gies (MRC and RANDOM) and five different RAM
reduction factors (
-x
and
-r
options respectively).
In the recomputation version used for the evaluation,
both the Largest subtree first and the Priority List op-
timizations were activated. All experiments were ex-
ecuted on a 48-core AMD system with 256GB RAM.
For all runs, RAM memory usage was measured ev-
ery 600 seconds with
top
.
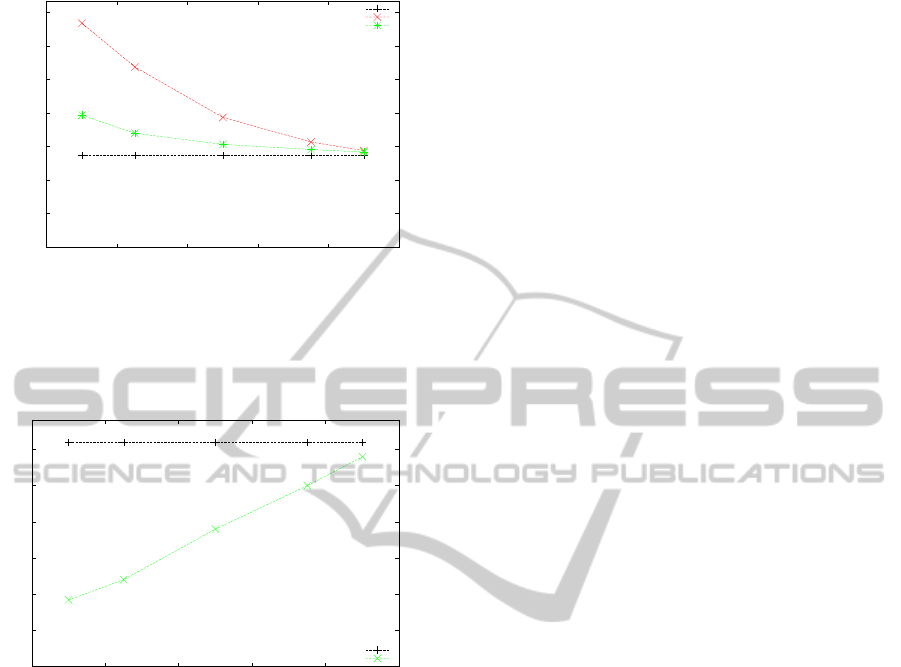
Figure 4 shows the corresponding decrease in
RAM usage. Values in Figure 4 correspond to maxi-
mum observed RAM usage values.
Figure 3 depicts the run time increase as a func-
tion of available space for storing ancestral proba-
bility vectors. Clearly, the MRC strategy outper-
forms the RANDOM strategy and the induced run-
time overhead, even for a reduction of available RAM
space to only 10% is surprisingly small (approxi-
mately 40%). This slowdown is acceptable, consid-
ering that instead of analyzing a large dataset on a
machine with 256GB, a significantly smaller and less
expensive system with, for instance, 32GB RAM will
be sufficient. Given the checkpointing capabilities of
RAxML-Light, the increase in run times does not rep-
resent a problem.
4.2 Evaluation of Traversal Overhead
In order to evaluate the overhead of the extended
(larger) tree traversals due to the required additional
ancestral probability vector computations, we modi-
fied the source code to count the number of ancestral
vector computations. We distinguish between three
cases with different recomputation costs (see Sec-
tion 3). For each case, there exists a dedicated PLF
implementation in RAxML.
Tip/Tip Both child nodes are tips.
Tip/Vector One child node is a tip, and the other is
an ancestral vector (subtree).
Vector/Vector Both child nodes are ancestral vec-
tors.
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS
93
0
2000
4000
6000
8000
10000
12000
14000
0 0.2 0.4 0.6 0.8 1
Average Running time (s)
Reduction Factor (% inner vectors on RAM)
Strategy performance for 10 runs of dataset 1500
std
recomRANDOM
recomSLOT_MIN_COST
Figure 3: Different replacement strategies. The dataset was
run with RAM allocations of 10%, 25%, 50% , 75%, and
90%, of the total required memory for storing all probability
vectors. Run times are averaged across 10 searches with
distinct starting trees.
0
5
10
15
20
25
30
0 0.2 0.4 0.6 0.8 1
Average Memory used (MB)
Reduction Factor (% inner vectors on RAM)
Memory measured for dataset 1500 for 10 trees
std
recomSLOT_MIN_COST
Figure 4: Overall RAM usage when allocating only 10%,
25%, 50% , 75%, and 90%, of the required ancestral proba-
bility vectors.
Table 1 shows a dramatic, yet desired increase,
in the number of Tip/Tip vector computations for the
MRC strategy. However, the amount of the slowest
type of ancestral node computations [Vector/Vector]
is only increased by 0.16% compared to the standard
implementation.
4.3 Evaluation of Full Tree Traversals
We created a simple test case that parses an input tree
and conducts 20 full tree traversals on the given tree.
This dedicated test code is also available at
https:
//github.com/fizquierdo/vectorRecomp/
. We
used the aforementioned starting trees and the 500,
1500, and 5000 taxon datasets. Each run was repeated
5 times and we averaged running times. All runs re-
turned exactly the same likelihood scores.
Table 2 indicates that, even for very small R values
(fraction of inner vectors allocated in memory) the run
time overhead is negligible compared to the standard
implementation.
5 CONCLUSIONS AND FUTURE
WORK
We have presented a generic strategy for the exact
computation of log likelihood scores and ML tree
searches with significantly reduced memory require-
ments. The additional computational cost incurred
by the larger number of required ancestral vector re-
computations is comparatively low when an appro-
priate vector replacement strategy is deployed. This
will allow for computing the PLF on larger datasets
than ever before, especially when the limiting factor is
available RAM. In fact, the memory versus additional
computationstrade-off can be adapted by the users via
a command line switch to fit their computational re-
sources. We also show that, the minimum number of
ancestral probability vectors for computing the PLF
that need to be kept in memory for a tree with n taxa
is log
2
(n) + 2. This result may be particularly inter-
esting for designing equally fast, but highly memory-
efficient phylogenetic post-analysis tools that rely on
full tree traversals.
Furthermore, the concepts presented here can be
applied to all PLF-based programs such as GARLI,
PHYML, MrBayes, etc. On the software engineering
side, the implementation of the recomputation strat-
egy in RAxML is encapsulated in such a way, that
it can be combined with any other memory saving
techniques such as out-of-core computations, Subtree
Equality Vectors, or the use of lossless compression
algorithms for ancestral probability vectors.
We plan to further refine/tune the MRC strategy
and fully integrate it into the up-to-date release of
RAxML-Light v1.0.5 and the already existing mem-
ory saving techniques. This will allow to infer trees
and compute likelihood scores on huge datasets, that
would previously have required a supercomputer, on
a single multi-core system.
ACKNOWLEDGEMENTS
Fernando Izquierdo-Carrasco is funded by the Ger-
man Science Foundation (DFG). The authors would
like to thank Simon Berger for pointing out and dis-
cussing favorable trade-offs regarding recomputation
of vectors and the potential use of lossless compres-
sion algorithms to decrease memory requirements.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
94

Table 1: Frequency of ancestral vector cases for the standard implementation and the recomputation strategies (50% of
ancestral vectors allocated).
Strategy Tip/Tip Tip/Vector Vector/Vector Total Runtime (s)
Standard 11,443,484 57,884,490 76,325,233 145,653,207 5678
MRC (0.5) 20,368,957 61,224,562 76,444,874 158,038,393 6453
Random (0.5) 37,778,575 85,303,730 104,398,910 227,481,215 7999
Table 2: Average run times in seconds for 20 full traversals averaged across 5 runs.
Dataset Standard R:=0.1 R:=0.9
500 0.122 0.121 0.130
1500 0.430 0.430 0.434
5000 2.402 2.412 2.438
REFERENCES
Barkan, E., Biham, E., and Shamir, A. (2006). Rigorous
bounds on cryptanalytic time/memory tradeoffs. In In
Advances in CryptologyCRYPTO 2006, volume 4117
of LNCS, pages 1–21. Springer-Verlag.
Berger, S. and Stamatakis, A. (2010). Accuracy and
performance of single versus double precision arith-
metics for Maximum Likelihood Phylogeny Recon-
struction. Springer Lecture Notes in Computer Sci-
ence, 6068:270–279.
Dri, P. and Galil, Z. (1984). A time-space tradeoff for
language recognition. Theory of Computing Systems,
17:3–12. 10.1007/BF01744430.
Felsenstein, J. (1981). Evolutionary trees from DNA se-
quences: a maximum likelihood approach. J. Mol.
Evol., 17:368–376.
Fletcher, W. and Yang, Z. (2009). INDELible: a flexible
simulator of biological sequence evolution. Molecular
biology and evolution, 26(8):1879–1888.
Guindon, S., Dufayard, J., Lefort, V., Anisimova, M.,
Hordijk, W., and Gascuel, O. (2010). New algorithms
and methods to estimate maximum-likelihood phylo-
genies: assessing the performance of PhyML 3.0. Sys-
tematic biology, 59(3):307.
Izquierdo-Carrasco, F. and Stamatakis, A. (2011). Comput-
ing the phylogenetic likelihood function out-of-core.
In In Proceedings of the IPDPS-HiCOMB 2011.
Martin Simonsen, T. M. and Pedersen, C. N. S. (2010).
Building very large neighbour-joining trees. Proceed-
ings of Bioinformatics 2010, to appear in Springer
bioinformatics lecture notes.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum
likelihood-based phylogenetic analyses with thou-
sands of taxa and mixed models. Bioinformatics,
22(21):2688–2690.
Stamatakis, A. (2011). Phylogenetic Search Algorithms for
Maximum Likelihood, pages 547–577. John Wiley &
Sons, Inc.
Stamatakis, A. and Alachiotis, N. (2010). Time and mem-
ory efficient likelihood-based tree searches on phy-
logenomic alignments with missing data. Bioinfor-
matics, 26(12):i132.
Stamatakis, A. and Ott, M. (2008). Exploiting fine-grained
parallelism in the phylogenetic likelihood function
with mpi, pthreads, and openmp: a performance study.
Pattern Recognition in Bioinformatics, pages 424–
435.
Vitter, J. S. (2008). Algorithms and data structures for ex-
ternal memory. Found. Trends Theor. Comput. Sci.,
2(4):305–474.
Wheeler, T. (2009). Large-scale neighbor-joining with
ninja. In Salzberg, S. and Warnow, T., editors, Al-
gorithms in Bioinformatics, volume 5724 of Lecture
Notes in Computer Science, pages 375–389. Springer
Berlin / Heidelberg.
Xu, J. and Lipton, R. J. (2005). On fundamental tradeoffs
between delay bounds and computational complexity
in packet scheduling algorithms. IEEE/ACM Trans.
Netw., 13:15–28.
Yang, Z. (1994). Maximum likelihood phylogenetic esti-
mation from DNA sequences with variable rates over
sites. J. Mol. Evol., 39:306–314.
Zwickl, D. (2006). Genetic Algorithm Approaches for the
Phylogenetic Analysis of Large Biological Sequence
Datasets under the Maximum Likelihood Criterion.
PhD thesis, University of Texas at Austin.
TRADING RUNNING TIME FOR MEMORY IN PHYLOGENETIC LIKELIHOOD COMPUTATIONS
95
