
Current Challenges on Polyp Detection in Colonoscopy
Videos: From Region Segmentation to Region
Classification. A Pattern Recognition-based Approach
Jorge Bernal, Javier S
´
anchez and Fernando Vilari
˜
no
Computer Vision Center and Computer Science Department UAB
Campus UAB, Edifici O, 08193, Bellaterra, Barcelona, Spain
Abstract. In this paper we present our approach on selection of regions of inter-
est in colonoscopy videos, which consists of three stages: Region Segmentation,
Region Description and Region Classification, focusing on the Region Segmenta-
tion stage. As part of our segmentation scheme, we introduce our region merging
algorithm that takes into account our model of appearance of the polyp. As the
results show, the output of this stage reduces the number of final regions and
indicates the degree of information of these regions. Our approach appears to
outperform state-of-the-art methods. Our results can be used to identify polyp-
containing regions in the later stages.
1 Introduction
Colon cancer, with an approximate number of 655.000 deaths worldwide per year, has
become the fourth leading cause of death by cancer in the United States and the third
leading cause in the Western world [1]. Colon cancer includes cancerous growths in the
colon, rectum and appendix. Colon cancer arises from adenomatous polyps in the colon
which can be identified by its prominent (flat or peduncular) shape.
Colon cancer has four distinct stages along with a fifth stage that is called ’recur-
ring’. Its survival rate (measured in five-year survival rate) decreases according to the
higher the stage the polyps are detected on, going from a 95% in stage I to a 3% in stage
IV [2], hence its importance to be detected on its early stages. There are several types
of screening techniques grouped according to the principles of functioning (depending
on if they need the on-line intervention of a physician or consist of introducing some
particle into the patient). One of the most used is colonoscopy. Colonoscopy [3] is a
procedure used to see inside the colon and rectum and it is able to detect inflamed tis-
sue, ulcers, and abnormal growths [4]. During colonoscopy, patients lie on their left side
on an examination table. The phyisician inserts a long and flexible tube called colono-
scope into the anus and guides it slowly through the rectum and into the colon. A small
camera is mounted on the scope and transmits a video image from inside the large intes-
tine to a computer screen, allowing the doctor to examine carefully the intestinal lining.
During this process the physician can remove polyps and later test them in a laboratory
to look for signs of cancer. Colonoscopy allows a direct visualization of the intestinal
Bernal J., Sánchez J. and Vilariño F..
Current Challenges on Polyp Detection in Colonoscopy Videos: From Region Segmentation to Region Classification. A Pattern Recognition-based
Approach.
DOI: 10.5220/0003352000620071
In Proceedings of the 2nd International Workshop on Medical Image Analysis and Description for Diagnosis Systems (MIAD-2011), pages 62-71
ISBN: 978-989-8425-38-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
surface but it has some drawbacks, such as the risk of perforation, the intervention cost,
or visualization difficulties among others.
The global objective of our project is to develop a tool that can indicate the doctor
which areas of the colon are more likely to contain a polyp by means of computer vision
techniques. The results of this tool can be used in several applications such as: 1) real-
time polyp detection, 2) off-line quality assessment of the colonoscopy, 3) quantitative
assessment of the trainee skills in training procedures, only to mention a few.
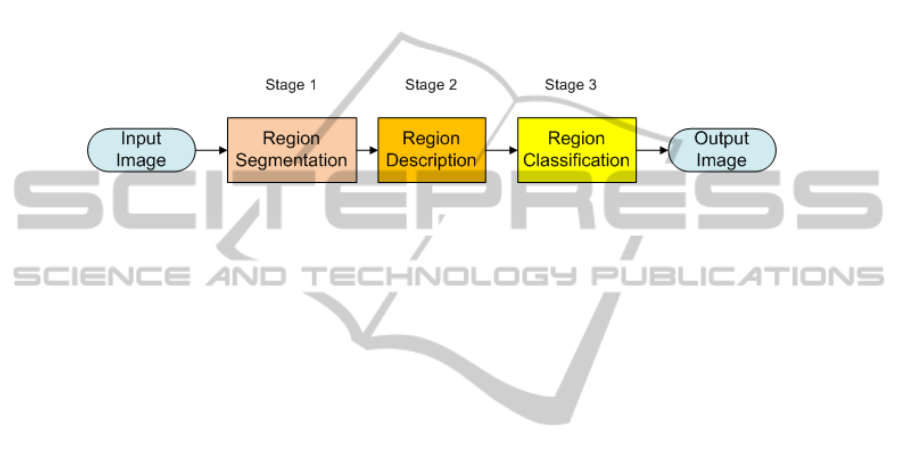
In order to achieve this goal we base our approach on a common Pattern Recognition
scheme (see Figure 1). The input to the scheme is a frame from a colonoscopy video
and the output will be this same image annotated in terms of polyp detection.
Fig. 1. General scheme of our approach.
The first stage, Region Segmentation, consists of segmenting automatically the im-
age in order to end up with a reduced number of relevant regions, one of them containing
a polyp. So the objectives are twofold: reduce the number of regions that should be ana-
lyzed and indicate which are the non-informative regions on where not even the human
eye can distinguish anything and therefore there will be no need to analyze them later.
The second and third stages (Region Description and Classification) are closely
related one to the other. Once we have a few regions from the segmentation stage, we
need to find which features in these regions can denote the presence or not of a cancer
polyp. The objective of the second stage will be to find a series of descriptors that
describe better what is a polyp region and what is not and, in the third stage, using the
knowledge acquired from the examples, classify the input regions into polyp-containing
candidates or not.
In order to develop and test our approach we rely on a database of thousands of
images extracted from 15 different videos of colonoscopy interventions.
The structure of this paper is as follows: In Section 2 we present our Region Seg-
mentation stage. In Section 3 we introduce the Region Description and Region Classi-
fication Stages. In Section 4 we present our preliminary experimental results. Finally,
in Section 5 we expose the main conclusions that have been extracted from this work
along with a presentation of the Future Work that we plan to do.
2 Region Segmentation
2.1 Our Model of Appearance of a Polyp
As we have mentioned before, we are dealing with colonoscopy images obtained from
real interventions videos. While observing the videos, we have found out that the light-
ing of the probe can give us hints about what is a polyp in an image. As the light
63
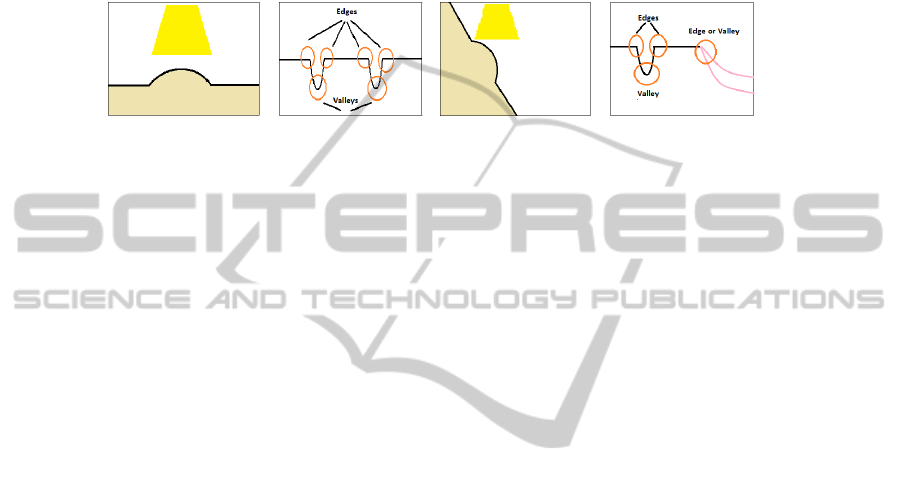
falls perpendicularly to the walls of the colon, it creates shadows around the surfaces
at which it is. More precisely, when the light falls into a prominent surface, it creates
a bright spot (with high grey-scale value) surrounded by darker areas, which are the
shadows, generating edges and valleys in the intensity image. This can be better under-
stood by looking at Figure 2, where we show an indication of the effect of light in the
intensity profiles, that depend on how the polyp appears (cenital or lateral view).
(a) (b) (c) (d)
Fig. 2. (a-b) Simulation of an overhead prominent surface and its correspondent grey-scale profile
(c-d) Simulation of a lateral prominent surface and its correspondent grey-scale profile.
Even considering this evidences of prominent surface appearance, there are some
challenges that need to be overcome:
– Non-uniform Polyp Appearance: first, the appearance of the polyp by itself is not
uniform, going from peduncular to flat shapes. Second, in most of the images we
will not have a clear vision of the polyp, viewing them from an overhead or lateral
views, which makes difficult a shape-based recognition scheme work.
– Uniform Colour Pattern: as all the tissues in the image present a very similar color
distribution, a segmentation scheme based purely on color will have difficulties to
segment correctly the image.
– Effect of the Reflections, which makes segmentation algorithms create artificial
regions in the image.
– Over and under Segmentation: We can have two problems related to the number
of segmented regions. Oversegmentation is related to having a large number of
very small regions, which implies a high computation cost to analyze them all) and
under-segmentation to the fact of having a smaller number of bigger regions, but
still higher than the number of structures that a human could identify on the image).
Taking these considerations into account, we base our segmentation method on a
model of appearance of a polyp that we have defined as a prominent shape enclosed
in a region that can be identified by the presence of edges and valleys. But we have to
take this as an indication, and we also should try to overcome the challenges we have
presented.
2.2 Region Segmentation Algorithm
In this subsection first we present the basics of each step of our segmentation approach
and at the end we show in Figure 4, step by step, a complete graphical example.
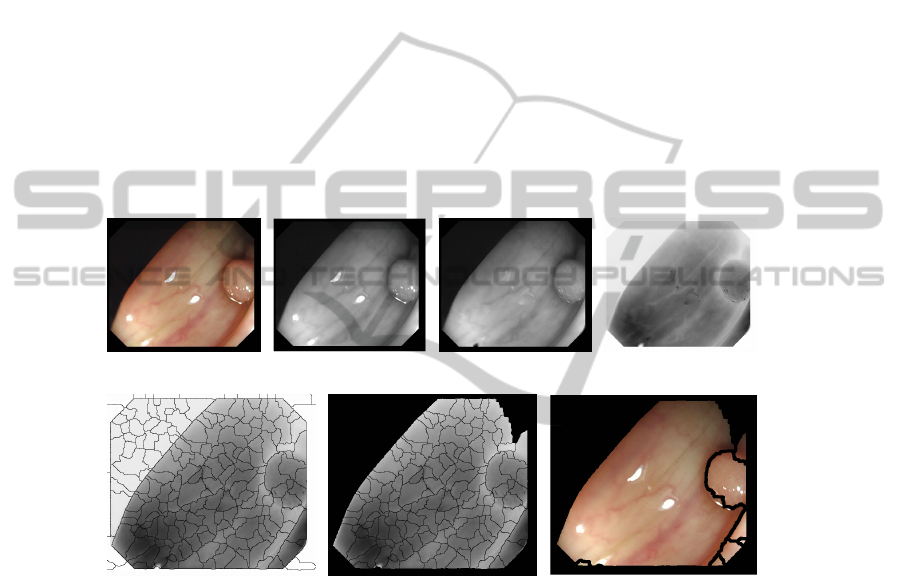
1. Image Preprocessing: Before applying any segmentation algorithm there are some
operations that should be done to the input image (Figure 4 a) in order to overcome
64
some of the challenges that were presented before. These preprocessing operations in-
clude: converting to gray-scale (Figure 4 b), image deinterleaving (as our images come
from a high definition interleaved video), correction of the reflections (Figure 4 c) and
obtaining the complemented version of the image (Figure 4 d).
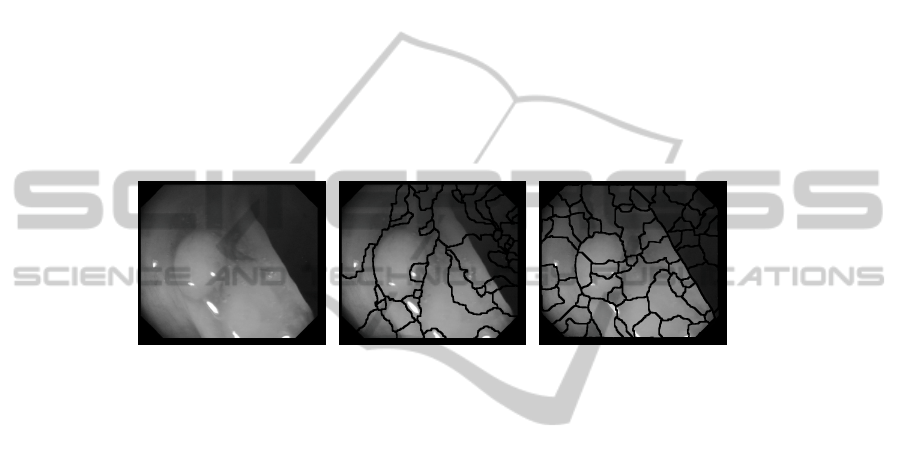
2. Segmentation: In this step we have several alternatives. We can use either simpler
(in terms of computation cost) methods such as watersheds [5] or go with algorithms
that are more powerful in terms of segmentation such as Mean-shift or Normalized
Cuts. We use watersheds to reduce the computation cost and also because more complex
approaches are generally color-based which do not seem so useful in our case. Another
point of our approach is that, instead of using the preprocessed image obtained in the
first step, we use gradient information, which, as it can be seen in Figure 3, encloses
better the structure of the shapes that appear on the image. After this step our image
will be divided in a large number of regions (Figure 4 e) that we will reduce by merging
them.
(a) (b) (c)
Fig. 3. (a) Original image (b) Original image segmentation (c) Gradient image segmentation.
3. Frontier-based Region Merging: in this step we focus on merging small neighbor
regions that are separated by weak frontiers (Figure 4 f). We denote as weak those
frontiers which present a low value of the weakness function (1).
F rontierW eakness = α ∗ edginess + β ∗ valleyness
+ γ ∗ anisotropic + η ∗ orientation
(1)
This weakness function is calculated using the response of the image to four different
weakness criteria: edges (by using a Canny detector the the image), valleyness (using a
ridges and valleys detector [6]), anisotropic filtering and gradient-based orientation. For
each one of the criteria we create a binary mask and take as first measure the percentage
of pixels that fall under the dark side of the mask. Once we have this percentage for each
of the criteria, we can decide if a frontier is weak according to a weakness criteria by
setting up a threshold.
We merge regions until there are no weak frontiers between regions (up to a thresh-
old value of the weakness function) or until the number of regions is stabilized.
4. Region-based Region Merging: in this step, as it did not happen in the previous
one, we consider when merging not only the weakness of the frontiers (by using a
different weakness function (2)) but also the grey-level content of the regions. In this
65
case the weakness measure takes into account which frontiers are kept after applying
consecutively two order-increasing median filters and a botton-hat mask.
F rontierW eakness = µ ∗ median + ρ ∗ bottom − hat (2)
Then we categorize the regions and frontiers, in terms of the amount of information
that they contain (i.e., low information means a region with a very high or very dark
mean grey level and very low standard deviation) and then merge compatible (same
kind of information) regions that have compatible weak frontiers. The objective here is
to end up with a reduced number of large regions as uniform as possible.
We merge regions until there are no weak frontiers between compatible regions (up
to a threshold weakness value) or until the number of regions is stabilized.
The objective of our segmentation and region merging sub-steps is twofold: first ob-
taining a good segmentation of the image that fits the structure of the several objects that
appear in it, such as polyps, and second, join and label those parts of the image where
we know we will not find a polyp inside and therefore, we should not process them in
order to save resources. We show in Figure 4 one complete segmentation process.
(a) (b) (c) (d)
(e) (f) (g)
Fig. 4. (a) Original image (b) Grey-scale (c) Reflection corrected (d) Preprocessed image (e) First
segmentation (184 regions) (f) Before region-based merging (136 regions) (g) Final image (9
regions).
3 Region Description and Region Classification
The objective of the Region Description stage is to choose a series of descriptors that
represent better what is a polyp region and what is not. We have made an study of the
bibliography of Feature Descriptors [7], dividing them into four groups: Shape Descrip-
tors, Color Descriptors, Texture Descriptors and Motion Descriptors. Our approach to
this stage is not to rely on one only type of descriptors and, as possible, try to use really
informative descriptors.
66
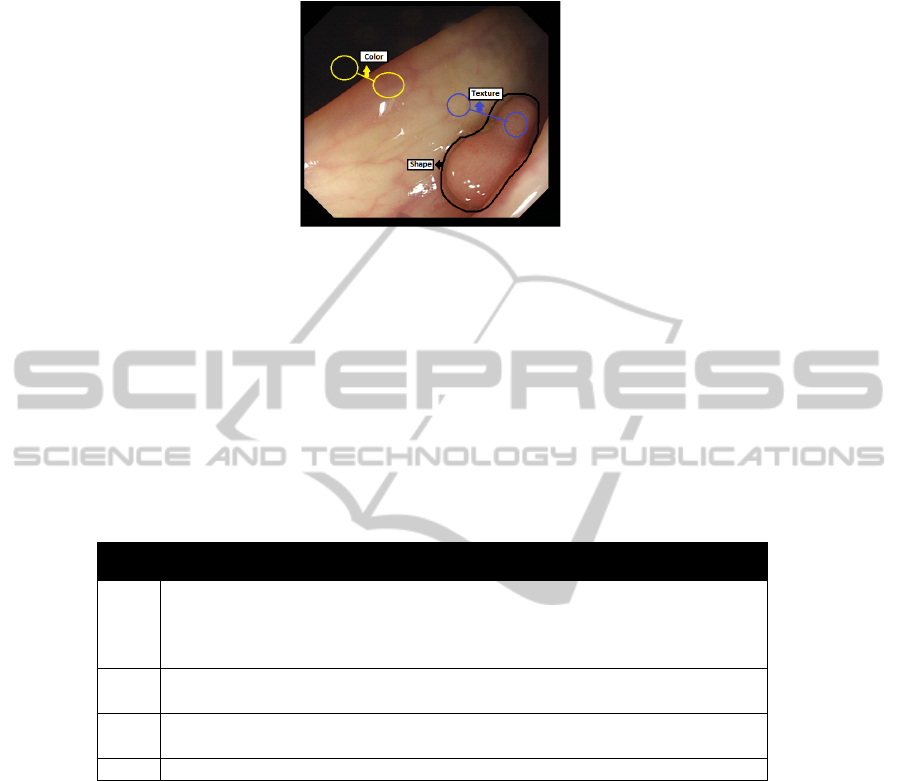
Fig. 5. Example of the need to use several types of descriptors.
If we take a look at the example, we can see that each of the types of descriptors
may have a role in our system. For example, we can see that the polyp is enclosed by a
closed contour and could be approximated by an ellipsoidal shape, so here we could use
Shape Descriptors. Color as it can be seen can be an important cue when defining what
is clearly not a polyp and we can also observe the difference of Texture between the
polyp (more granular) and non-polyp regions (more plain). In Table 1 we show some of
the most important ones of each group.
Table 1. Examples of several feature descriptors.
Type Methods
Shape Contour-based: Global (Wavelets [8], Fourier [9], Shape Signature [10]) or Struc-
tural (Chain Code [11], Blurred Shape Model [12], Shape Context [13]).
Region-based: Global (Angular Radial Partitioning [14], Zernike Moments [15],
Shape Matrix [16]) or Structural (Skeletons).
Color Scalable Color Descriptor [17], Color Structure Descriptor [18], Color Constant
Color Indexing [19].
Texture SIFT [20], SURF [21], Texture Browsing Descriptor [22], Local Binary Patterns
[23], Co-ocurrence Matrices [24].
Motion Optical Flow [25], Angular Circular Motion [26].
Once we have described our regions we can start classifying them. As happens with
the Region Description step, this stage has not been implemented yet in our method be-
cause of two reasons: first, time constraints (since we just started the development of our
approach) and secondly, and more important, because we have a strong belief in that the
classification system is good as long as its inputs (outputs from the previous stages) are
good. As in many Pattern Recognition-based methods, we will use a machine learning
approach [27]. A learner can take advantage of examples (in our case, descriptions of
polyp and non-polyp containing regions) in order to capture characteristics of interest
of their unknown underlying probability distribution.
We will provide the system examples of regions that contain polyps and examples
of regions that no contain polyps. These example regions will be described and incor-
porated into the machine learning algorithm of choice in order to learn a polyp and
67

non-polyp pattern. New input images will be automatically segmented, described and
incorporated into the testing step with the objective of finding out if they are more near
to be a polyp candidate region or non-polyp candidate region. In our case our success
will be measured not only on terms of how many true positives we get but also on the
number of false negatives. It seems clear that it is harmless to identify one non-polyp
region as a polyp region that the opposite.
4 Experimental Results
4.1 Experimental Setup
In order to test the performance of our segmentation algorithm (because we have only
implemented the Region Segmentation stage until now) we have created a database
which consists of 150 different studies of polyp appearance in colonoscopy videos.
We have also a set of masks for each image (one that indicates the polyp position for
each image and another that indicates the non-informative regions) that will be used to
calculate the different performance measures.
We will use two different performance measures: Annotated Area Covered (AAC)
and Dice Similarity Coefficient (DICE). The first one indicates the maximum percent-
age of polyp that is contained on a single region while the second indicates, for the
region that covered more the polyp, the percentage of polyp information out of all the
region information. We will compare our method with Normalized Cuts [28] for both
polyp detection and non-informative region characterization.
4.2 Experimental Results
We show in Table 2 experimental results for both our method and Level Sets. The results
show that our approach is better than Normalized Cuts in terms of AAC but it is worse
than Normalized Cuts in terms of DICE, although our results for peduncular studies are
similar than the ones achieved by Normalized Cuts.
Table 2. Summary of the results.
Our Method Normalized Cuts
Measure /
Database
Whole Flat Peduncular Whole Flat Peduncular
Polyp AAC 96.5% 97.96% 92.79% 76.88% 79.33% 70.75%
Polyp DICE 22.07% 14% 42.23% 35.38% 28.10% 51.67%
Non-
informative
96.86% 96.46% 97.88% 80.38% 77.83% 86.71%
The reason for the great difference in DICE results lie on the fact that our method
divides the polyp area into smaller regions while Normalized Cuts fits the polyp area in
a smaller number of regions. So in order to improve our results we should find a way
to merge small polyp regions into bigger ones. We get better results than Normalized
Cuts in terms of AAC because our regions are smaller so they contain less both polyp
and non-polyp information and, in the case of a polyp region, as they are smaller, they
68
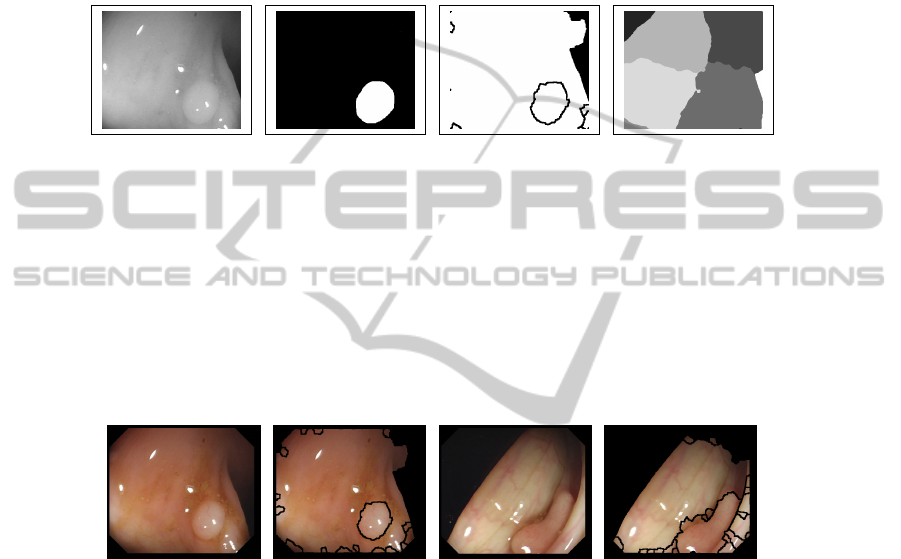
contain more polyp information. We show in Figure 6 qualitative segmentation results
(which have been calculated, for the sake of comparison, not taking into account the
black borders of the image). In general, the regions segmented by our method fit better
the structure of the polyp but as they are smaller, they can miss some of the polyp
information. Normalized Cuts regions, as they are bigger, cover more polyp information
but they do not approximate well the shape of the polyp. Our method also gives better
results in terms of detecting which regions of the image are non-informative.
(a) (b) (c) (d)
Fig. 6. (a) Original images (b) Polyp mask (c) Final regions with our method (c) Final regions
with Normalized Cuts.
So, taking into account that Region Segmentation is only the first stage of our ap-
proach (and that it is not finished yet) it seems clear that, if we are going to describe
later our regions to decide if they contain a polyp or not, our method is more suitable
than Normalized Cuts because our regions approximate better the polyp. To finish this
section we show in Figure 7 we show some preliminary qualitative segmentation re-
sults, that show that first, we are reducing the search area of the image by eliminating
non-informative regions and second, that we end up with a low number of regions.
(a) (b) (c) (d)
Fig. 7. (a-c) Original images (b-d) Segmented images.
5 Conclusions
Our objective is to detect polyps in colonoscopy videos. To do so, we propose a Pattern
Recognition scheme divided in three main stages. The first one, Region Segmentation,
is done with two objectives: reduce the size of the problem (as one of our possible
applications can be real-time polyp detection) as much as possible, and provide as result
to the later stages of the process chain a set of regions of interest. In order to segment
the image we have studied in depth the structure of several polyp-containing images
from our video database, with the aim of finding cues that can let us discern which
regions have some evidence of containing polyps and which not (in this case, we will
not analyze again these regions). So far, our method reduces greatly the number of
69

regions, offering as output a small number of them that, in some cases, cover the whole
shape of the polyp in just one region and, as the experimental results show, we identify
well the non-informative regions of the image.
Once we have this subset of regions of interest, the next step, Region Description,
will consist of describing them in terms of features to characterize them. This step will
be the seed to the final stage, Region Classification, where our plan is to implement
a machine learning approach. The ideal output will be a mask superimposed to the
image that indicates the region of the image that more likely contains a polyp. The
segmentation results give us hope that, after some improvements, we will be able to
start with the description step.
Acknowledgements
The authors would like to thank Dr. Antonio L
´
opez and Dr. Debora Gil for their helpful
comments and suggestions. This work was supported in part by a research grant from
Universitat Auton
`
oma de Barcelona 471-01-3/08, by the Spanish Government through
the founded project ”COLON-QA” (TIN2009-10435) and by research programme Con-
solider Ingenio 2010: MIPRCV (CSD2007-00018).
References
1. National Cancer Institute, “What You Need To Know About Cancer of the Colon and Rec-
tum,” U.S. National Institutes of Health, pp. 1–8, 2010.
2. A. Tresca, “The Stages of Colon and Rectal Cancer,” New York Times (About.com), p. 1,
2010.
3. J. Hassinger, S. Holubar, et al., “Effectiveness of a Multimedia-Based Educational Interven-
tion for Improving Colon Cancer Literacy in Screening Colonoscopy Patients,” Diseases of
the Colon & Rectum, vol. 53, no. 9, p. 1301, 2010.
4. National Digestive Diseases Information Clearinghouse, “Colonoscopy,” National Institute
of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH, p. 1, 2010.
5. L. Vincent and P. Soille, “Watersheds in digital spaces: an efficient algorithm based on
immersion simulations,” IEEE Transactions on Pattern Analysis and Machine Intelligence,
vol. 13, no. 6, pp. 583–598, 1991.
6. A. L
´
opez, F. Lumbreras, et al., “Evaluation of methods for ridge and valley detection,” IEEE
Transactions on Pattern Analysis and Machine Intelligence, vol. 21, no. 4, pp. 327–335,
1999.
7. J. Bernal, F. Vilari
˜
no, and J. S
´
anchez, “Feature Detectors and Feature Descriptors: Where
Are Now,” Tech. Rep. 154, Computer Vision Center, Universitat Aut
`
onoma de Barcelona,
September 2010.
8. C. H. Chuang and C. C. J. Kuo, “Wavelet descriptor of planar curves: Theory and applica-
tions,” IEEE Transactions on Image Processing, vol. 5, no. 1, pp. 56–70, 2002.
9. H. Kauppinen, T. Seppanen, and M. Pietikainen, “An experimental comparison of autore-
gressive and Fourier-based descriptors in 2D shape classification,” IEEE Transactions on
Pattern Analysis and Machine Intelligence, p. 201, 1995.
10. D. Zhang and G.Lu, “Review of Shape Representation and Description Techniques,” Pattern
Recognition, vol. 37, no. 1, pp. 1–19, 2004.
70

11. J. Sun and X. Wu, “Shape Retrieval Based on the Relativity of Chain Codes,” Lecture Notes
in Computer Science, Multimedia Content Analysis and Mining, vol. 4577/2007, pp. 76–84,
2007.
12. A. Forn
´
es and S. E. et al., “Handwritten Symbol Recognition by a Boosted Blurred Shape
Model with Error Correction,” Pattern Recognition and Image Analysis, pp. 13–21, 2007.
13. J. Bohg and D. Kragic, “Grasping Familiar Objects Using Shape Context,” in International
Conference on Advanced Robotics. ICAR 2009, pp. 1–6, June 2009.
14. A. Chalechale, A. Mertins, and G. Naghdy, “Edge image description using angular radial
partitioning,” IEEE Proceedings-Vision Image and Signal Processing, vol. 151, no. 2, pp. 93–
101, 2004.
15. A. Khotanzad and Y. H. Hong, “Invariant Image Recognition by Zernike Moments,” IEEE
Transactions on Pattern Analysis and Machine Intelligence, vol. 12, pp. 489–497, 1990.
16. D. Zhang and M. C. Y. Lim, “An efficient and robust technique for region based shape
representation and retrieval,” in 6th IEEE/ACIS International Conference on Computer and
Information Science, 2007. ICIS 2007, pp. 801–806, 2007.
17. D. Borghesani, C. Grana, and R. Cucchiara, “Color Features Performance Comparison for
Image Retrieval,” Image Analysis and Processing–ICIAP 2009, pp. 902–910, 2009.
18. M. Kundu, M. Banerjee, and P. Bagrecha, “Interactive Image Retrieval in a Fuzzy Frame-
work,” Fuzzy Logic and Applications, vol. 5571, pp. 246–253, 2009.
19. B. V. Funt and G. D. Finlayson, “Color Constant Color Indexing,” IEEE Transactions on
Pattern Analysis and Machine Intelligence, vol. 17, no. 5, pp. 522–529, 1995.
20. D. G. Lowe, “Object Recognition from Local Scale-Invariant Features,” in International Con-
ference on Computer Vision, Corfu, Greece, pp. 1150–1157, Published by the IEEE Com-
puter Society, 1999.
21. H. Bay, T. Tuytelaars, and L. V. Gool, “SURF: Speeded Up Robust Features,” Computer
Vision–ECCV 2006, pp. 404–417, 2006.
22. K. L. Lee and L. H. Chen, “An efficient computation method for the texture browsing de-
scriptor of MPEG-7,” Image and Vision Computing, vol. 23, no. 5, pp. 479–489, 2005.
23. T. Ojala, M. Pietikainen, and T. Maenpa, “Gray scale and rotation invariant texture classifi-
cation with local binary patterns,” Computer Vision-ECCV 2000, pp. 404–420, 2000.
24. J. R. Carr and F. P. de Miranda, “The semivariogram in comparison to the co-occurrence
matrix for classification of image texture,” IEEE Transactions on Geoscience and Remote
Sensing, vol. 36, pp. 1945–1952, nov 1998.
25. B. K. P. Horn and B. G. Schunck, “Determining Optical Flow,” Artificial Intelligence,
vol. 17, no. 1-3, pp. 185–203, 1981.
26. B. Erol and F. Kossentini, “Local Motion Descriptors,” in IEEE Workshop on Multimedia
Signal Processing, vol. October 2001, pp. 467–472, 2001.
27. C. Bishop et al., Pattern recognition and machine learning. Springer New York, 2006.
28. J. Shi and J. Malik, “Normalized cuts and image segmentation,” Pattern Analysis and Ma-
chine Intelligence, IEEE Transactions on, vol. 22, no. 8, pp. 888–905, 2002.
71
