
CONVENTIONAL AND BAYESIAN VALIDATION
FOR FUZZY CLUSTERING ANALYSIS
Olfa Limam
LARODEC, ISG, University of Tunis, Tunis, Tunisia
Fouad Ben Abdelaziz
American University of Sharjah, Sharjah, U.A.E.
Keywords:
Bayesian validation, Fuzzy set, Fuzzy clustering methods.
Abstract:
Clustering analysis has been used for identifying similar objects and discovering distribution of patterns in
large data sets. While hard clustering assigns an object to only one cluster, fuzzy clustering assigns one object
to multiple clusters at the same time based on their degrees of membership. An important issue in clustering
analysis is the validation of fuzzy partitions. In this paper, we consider the Bayesian like validation along with
four conventional validity measures for two clustering algorithms namely, fuzzy c-means and fuzzy c-shell
based. An empirical study is conducted on five data sets to compare their performances. Results show that
the Bayesian validation score outperforms the conventional ones. However, a multiple objective approach is
needed.
1 INTRODUCTION
Fuzzy cluster analysis aims at dividing data into
groups or clusters such that items within a given clus-
ter have a high degree of similarity whereas items of
different groups have a high degree of dissimilarity
(Klawonn and Hoppne, 2009). Objects are not clas-
sified as belonging to one and only one cluster but
instead, they all possess a degree of membership for
each cluster. There are many fuzzy cluster analysis
techniques available and they have been successfully
applied to several data analysis problems (Klawonn
and Hoppne, 2009).
In this context, we conduct a comparative study
of the performance of two known fuzzy clustering al-
gorithms, Fuzzy c-means (FCM) and Fuzzy c-shell
method (FCS). Once the clustering algorithm is ap-
plied, the second step is to decide on the optimal
number of clusters using cluster validity measures.
For this evaluation, we use four conventional and
bayesian cluster validity indices (Carvalho, 2006).
This paper is organized as follows. Section 2 presents
a brief review of two clustering algorithms, namely
FCM and FCS. Section 3 reports the experimental
environment and comparison result and Section 4
presents a brief conclusion.
2 FUZZY CLUSTERING
ALGORITHMS
In this study, we present a comprehensive compar-
ative analysis using two fuzzy clustering algorithms
FCM and FCS. First, we explain their fundamental
concepts.
2.1 The Fuzzy C-Means Method
FCM reveals structure in data through minimizing
a quadratic objective function (Graves and Pedrycz,
2010). The formulation of FCM optimization model
is:
Minimize J(U,V) =
c
∑
i=1
n
∑
k=1
u
m
ik
d
2
(x
k
,v
i
) (1)
subject to the constraint
u
ik
∈ [0,1] (2)
c
∑
i=1
u
ik
= 1,∀k ∈ {1...n}, (3)
where n is the total number of patterns in a given
dataset, c is the number of clusters, X = {x
1
,x
2
,...,x
n
}
the feature data, V = {v
1
,...,v
c
} cluster centroids,
135
Limam O. and Ben Abdelaziz F..
CONVENTIONAL AND BAYESIAN VALIDATION FOR FUZZY CLUSTERING ANALYSIS.
DOI: 10.5220/0003110201350140
In Proceedings of the International Conference on Fuzzy Computation and 2nd International Conference on Neural Computation (ICFC-2010), pages
135-140
ISBN: 978-989-8425-32-4
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

U = [u
ik
]
c∗n
denotes the fuzzy partition matrix, u
ik
is
the membership degree of pattern x
k
to the ith cluster.
The distance d(x
k
,v
i
), noted as d
ik
, is the Euclidean
distance norm between object x
k
and center v
i
and m
is the fuzzification exponent. The respective mem-
bership functions and cluster centroids to solve the
constrained optimization problem, given in (Bezdek,
1974) are as follows:
v
i
=
∑
n
k=1
u
m
ik
∗ x
k
∑
n
k=1
u
m
ik
, (4)
and
u
ik
=
1
∑
c
j=1
(
d
2
ik
d
2
jk
)
1/m−1
. (5)
FCM algorithm is stated as follows:
1. Fix c, 2 ≤ c ≺ n, fix m, 1 ≤ m ≤ ∞, initialize the
fuzzy membership matrix U = [u
ik
]
c∗n
, .
2. Calculate c fuzzy cluster centers using Equation
4.
3. Update the membership matrix U using Equation
5, until ||U
l
−U
l−1
|| < ε, then stop.
4. Else return to 2.
FCM algorithm has proven to be a very popular
clustering method. Mainly, it is applied for hyper-
spherical shaped data. However, FCM has some
shortcomings. It suffers from the convergence to lo-
cal minimum when the number of clusters increases.
Also, it shows a sensitivity to initialization parame-
ter values and dependence on data characteristics. It
requires initialization for prototypes while actually a
good initialization is difficult to assess. Moreover, it
requires the number of clusters to be known a priori.
Therefore, an extensive evaluation is required (Wang
and Zhang, 2007).
2.2 The Fuzzy C-Shell Method
FCS is an extension of the FCM algorithm where the
boundaries of spheres and ellipsoids are detected. The
prototype for a circular shell cluster is described by its
center point and a shell radius as an additional param-
eter, v
i
, r
i
, respectively (Dave, 1990). The objective
function for FCS is given by :
Minimize J(U,v,r) =
c
∑
i=1
n
∑
k=1
u
m
ik
d(x
k
,(v
i
,r
i
))
2
, (6)
where the distance measure d(x
k
,(v
i
,r
i
)) is the Eu-
clidean norm between x
j
,v
i
and r
i
and it is defined as
follows:
d(x
k
,(v
i
,r
i
))
2
= (||x
k
− v
i
||) − r
i
)
2
. (7)
FCS algorithm produces a fuzzy partition of the data
set via optimization of the previous objective func-
tion. The resulting cluster centroids and membership
functions are given by the following:
v
i
=
∑
n
k=1
u
m
ik
∗ x
k
∑
n
k=1
u
m
ik
, (8)
u
ik
=
1
∑
c
j=1
(
d
2
ik
d
2
jk
)
2/m−1
. (9)
FCS algorithm is stated as follows:
1. Fix c, 2 ≤ c ≤ n, Fix m, initialize the fuzzy mem-
bership matrix U = [u
ik
]
c∗n
.
2. Calculate cluster centers using Equation 8 and up-
date memberships u
ij
based on Equation 9
3. Calculate the d
ik
defined in Equation 7.
4. Update the membership matrix, until ||U
l
−
U
l−1
|| < ε then stop
5. Else go to step 3.
FCS algorithms are computationally expensive be-
cause their updating equation requires a non linear
equation to be solved iteratively. Hence, it requires a
suitable method to solve non-linear equations (Dave,
1990). In the following section, we conduct a com-
parative study to assess the performance of different
cluster validity measures.
3 A COMPARATIVE STUDY OF
FUZZY CLUSTER ANALYSIS
First, we introduce conventional and bayesian valida-
tion cluster validity measures. Then, we conduct an
experimental study on five datasets.
3.1 Conventional and Bayesian Cluster
Validity Measures
There are many fuzzy clustering validity indices to
evaluate clustering results (Cho and Yoo, 2006). In
this section, we review five of them, namely partition
coefficient, classification entropy, Fukuyama-Suengo,
Xie-beni Index and Bayesian Score.
3.1.1 Partition coefficient
(Bezdek, 1974) proposed a cluster validity index for
fuzzy clustering named partition coefficient (PC).
This index determines the performance measure
ICFC 2010 - International Conference on Fuzzy Computation
136

based on minimizing the overall content of pairwise
fuzzy intersection in U. The PC index is given by:
PC(U,c) =
∑
n
j=1
∑
c
i=1
(u
ij
)
2
n
, (10)
ranging within [1/c,1]. Optimal number of clusters is
obtained by maximizing the value of PC with respect
to a certain value of c. However, this index does not
perform well in large datasets because the value of PC
decreases monotonically when n gets large (Halkidi
et al., 2001)(Cho and Yoo, 2006)
(Wang and Zhang, 2007).
3.1.2 Classification entropy
Also, (Bezdek, 1974) proposed the classification en-
tropy (CE). It is a scalar measure of the amount of
fuzziness in a givenU and it is one of the most widely
used cluster validity indices. CE index is defined as
follows :
CE(U,c) =
−
∑
n
j=1
∑
c
i=1
u
ij
log
a
(u
ij
)
n
, (11)
where, CE values range within [0,log
a
c], with a is the
base of logarithm. Optimal partition is obtained by
minimizing the value of CE with respect to a certain
value of c. Also, CE is monotonically decreasing as c
gets larger.
The PC and CE indices use only membership val-
ues and do not take into consideration of cluster struc-
ture (Halkidi et al., 2001)(Cho and Yoo, 2006)
(Wang and Zhang, 2007).
3.1.3 Fukuyama-Suengo
(Fukuyama and Suengo, 1989) proposed a
Fukuyama-Suengo (FS) to validate the cluster-
ing by combining the compactness and separateness.
The FS index is given by the following:
FS(U,V,X) =
c
∑
i=1
n
∑
j=1
(u
ij
)
m
(||x
j
− v
i
||
2
− ||v
i
− ¯v||
2
),
(12)
where ¯v =
∑
n
i=1
x
i
/n. The term ||x
j
− v
i
||
2
measures
the compactness of clusters as the distance between
the representation element x
j
and cluster centroids v
i
of each cluster i, and ||v
i
− ¯v||
2
measures the sepa-
ration between each cluster centroid v
i
and the mean
of cluster centroids ¯v. An optimal cluster is founded
by minimizing FS to produce the best fuzzy partition
result of a given dataset. FS, as PC and CE, is mono-
tonically decreasing when c gets large (Halkidi et al.,
2001)(Cho and Yoo, 2006)
(Wang and Zhang, 2007).
3.1.4 Xie-beni Index
(Xie and Beni, 1991) proposed a Xie-beni Index (XB)
as a validity index based on compactness and sepa-
rateness. The XB index is defined as follows:
XB(U,V,X) =
∑
c
i=1
∑
n
j=1
u
2
ij
(||v
i
− x
j
||
2
n∗ d
2
min
, (13)
where d
min
= min
i, j
||v
i
− v
j
||. The numerator mea-
sures the compactness of the fuzzy partition, using the
distance between the center of cluster v
i
and a repre-
sentative element x
j
, weighted by the fuzzy partition
membership of data point j to cluster i. The denomi-
nator, denoting the strength of the separation between
clusters is defined as the minimum distance between
cluster centers weighted by n. The good partition is
found by minimizing XB index with respect to a cer-
tain value of c. XB index is monotonically decreasing
when the number of clusters gets very large and close
to n (Halkidi et al., 2001)(Cho and Yoo, 2006)
(Wang and Zhang, 2007)(Yang et al., 2006).
3.1.5 Bayesian Score
(Cho and Yoo, 2006) proposed a Bayesian score (BS)
by formally transferring the principles of the clas-
sic Bayes’ theorem to memberships. Unlike conven-
tional measures, based on the distance between clus-
ters, the Bayesian like validation method selects the
fuzzy partition with the highest membership degree
in the dataset. It selects a partition with the maximum
membership as an optimal cluster partition. The BS
index is given in the following:
BS =
∑
c
i=1
P(C
i
/D
i
)
C
(14)
=
∑
c
i=1
Π
n
j=1
P(C
i
)P(d
ij
|C
i
)/P(d
ij
)
C
,
P(c
i
) and P(d
ij
) are calculated as follows:
P(C
i
) =
∑
n
j=1,u
ij
≥α
u
ij
∑
n
j=1
∑
c
i=1
u
ij
, (15)
P(d
ij
) =
c
∑
i=1
P(C
i
)P(d
ij
/C
i
) =
c
∑
i=1
P(C
i
)u
ij
, (16)
where D
i
= {d
ij
/u
ij
> α,1 ≤ j ≤ n} and N
i
= n(D
i
).
The optimal number of clusters is chosen where BS is
maximized (Halkidi et al., 2001)(Cho and Yoo, 2006).
All indices suffer from their monotonous depen-
dence on the number of clusters. They show their
sensitivity to the initialization parameter, more specif-
ically to the fuzzifier m, and they lack direct con-
nection to the dataset structure. Hence, they do not
CONVENTIONAL AND BAYESIAN VALIDATION FOR FUZZY CLUSTERING ANALYSIS
137
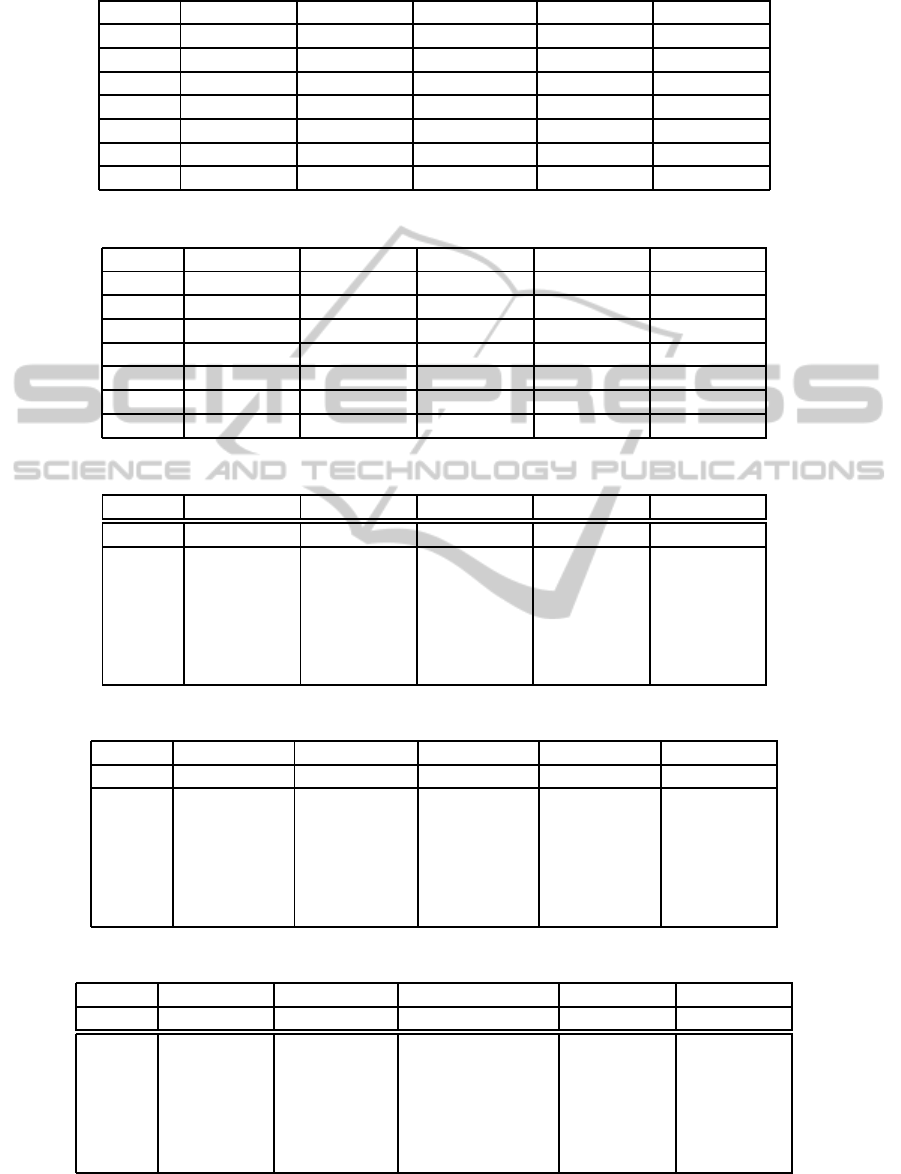
Table 1: Cluster validity values for Yeast dataset.
PC CE FS XB BS
Cluster FCM FCS FCM FCS FCM FCS FCM FCS FCM FCS
5 0.84 0.90 0.08 0.010 61.53 56.23 2.60 1.80 0.16 0.15
6 0.82 0.89 0.09 0.010 58.84 55.20 1.79 1.09 0.24 0.22
7 0.81 0.84 0.11 0.020 57.07 50.02 1.30 1.02 0.34 0.28
8 0.80 0.85 0.12 0.013 56.06 51.06 9.96 4.52 0.33 0.20
9 0.79 0.82 0.13 0.015 55.42 59.42 7.86 5.78 0.39 0.36
10 0.79 0.79 0.12 0.019 55.64 53.09 6.30 5.70 0.41 0.46
Table 2: Cluster validity values for Wisconsin breast cancer dataset.
PC CE FS XB BS
Cluster FCM FCS FCM FCS FCM FCS FCM FCS FCM FCS
2 0.69 0.69 7.87 7.00 -1.61 -2.60 3.53 4.01 1.00 1.00
3 0.68 0.69 1.87 1.90 -1.63 -3.89 2.91 3.02 0.99 0.40
4 0.69 0.69 1.65 1.90 -1.50 -0.98 2.79 2.24 0.99 0.99
5 0.68 0.68 7.81 7.65 -1.62 -2.64 4.95 5.65 0.98 0.48
6 0.67 0.68 1.06 0.99 -1.62 6.32 4.91 4.89 0.69 0.98
7 0.68 0.68 6.05 6.34 -1.52 -3.24 1.24 2.24 0.68 0.45
Table 3: Cluster validity values for Abalone dataset.
PC CE FS XB BS
Cluster FCM FCS FCM FCS FCM FCS FCM FCS FCM FCS
11 0.37 0.39 0.25 0.11 0.47 0.40 0.39 0.36 0.02 0.09
12 0.40 0.42 0.24 0.22 0.41 0.38 0.37 0.39 0.03 0.01
13 0.39 0.39 0.24 0.12 0.42 0.39 0.42 0.65 0.08 0.07
14 0.43 0.37 0.23 0.21 0.45 0.47 0.39 0.31 0.01 0.02
15 0.37 0.37 0.22 0.09 0.40 0.41 0.37 0.40 0.11 0.23
16 0.42 0.45 0.23 0.19 0.37 0.30 0.30 0.36 0.13 0.25
Table 4: Cluster validity values for arrhythmia dataset.
PC CE FS XB BS
Cluster FCM FCS FCM FCS FCM FCS FCM FCS FCM FCS
25 0.96 0.99 0.023 0.042 -3.12 -3.05 12.91 9.99 0.14 0.20
26 0.96 0.99 0.023 0.059 -2.75 -2.56 2.96 4.87 0.13 0.34
27 0.97 0.97 0.021 0.017 -3.02 -3.87 1.127 2.98 0.16 0.34
28 0.96 0.98 0.022 0.031 -3.05 -3.05 0.52 2.25 0.16 0.21
29 0.97 0.96 0.021 0.011 -3.01 -3.02 0.30 0.90 0.17 0.36
30 0.96 0.97 0.023 0.001 -3.22 -3.33 0.18 0.08 0.13 0.19
Table 5: Cluster validity values for Iris dataset.
PC CE FS XB BS
Cluster FCM FCS FCM FCS FCM FCS FCM FCS FCM FCS
2 0.97 0.98 0.005 0.014 -333,84 -338,52 0,73 0.50 0.52 0.88
3 0.98 0.99 0.011 0.016 -449,22 -129,62 0,16 0.47 0.47 0.43
4 0.96 0.99 0.019 0.015 -437,27 -471,60 0,07 0.42 0.42 0.31
5 0.97 0.98 0.015 0.020 -476,19 -149,13 0,04 0.47 0.47 0.69
6 0.96 0.98 0.027 0.009 -486,11 -640,31 0,03 0.42 0.42 0.64
7 0.93 0.99 0.026 0.006 -480,83 -462,31 0,02 0.43 0.43 0.80
ICFC 2010 - International Conference on Fuzzy Computation
138

use the dataset itself, except XB and FS involve the
dataset structure (Halkidi et al., 2001). In the next
section, we compare the performance of the above
mentioned indices in determining the true number of
clusters.
3.2 Experimental Results
To evaluate fuzzy partitions obtained from FCM and
FCS, cluster validity measures introduced previously
are compared using five data sets : Yeast, Breast Can-
cer, Abalone, Arrhythmia and Iris. Results of this
comparison are given in Tables from 1 to 5. All ex-
periments are repeated six times on each dataset by
increasing number of clusters. The number of clus-
ters ranging between intervals adaptively to the real
number of clusters (reported on the first column) and
the fuzziness parameter value is m = 1.2.
Yeast data consists of 1484 samples with nine fea-
ture values and ten classes. Table 1 shows results of
Yeast data for all validation methods when the num-
ber of clusters ranges from 5 to 10. While PC, CE, FS
and XB fail to identify the optimal number of Yeast
clusters, BS index correctly identifies it for both clus-
tering algorithms: FCM and FCS.
Wisconsin Breast Cancer data consists of 286
samples, where each pattern has nineteen features and
two clusters. Table 2 shows results of Wisconsin
Breast cancer dataset, where, CE, FS and XB fail to
recognize the optimum number of Yeast clusters, but
BS and PC indices correctly identify the optimal num-
ber of clusters for both clustering algorithms: FCM
and FCS.
Abalone dataset contains 4177 samples, where
each pattern has 279 features values, and sixteen clus-
ters. Results for Abalone dataset are given in Table 3.
They show that CE fails to identify the optimal num-
ber of Yeast clusters, while, XB correctly identifies
the optimam number except when the FCM algorithm
is applied. However, PC, FS and BS correctly iden-
tify the optimal number of clusters for both clustering
algorithms: FCM and FCS.
Arrhythmia dataset consists of 452 samples with 8
dimensional measurement spaces and 29 classes. Ta-
ble 4 shows results of Arrhythmia dataset. We notice
that FS and XB fail to identify the optimal number of
Yeast clusters. While PC and CE correctly identify
the optimal number except FCM algorithm is applied.
BS correctly identifies the optimal number of clusters
for both clustering algorithms.
Iris dataset contains 150 samples with four at-
tributes and has three classes. Table 5 gives results
for Iris dataset. We note that FS, XB and BS fail to
identify the optimal number of Yeast clusters. CE cor-
rectly identifies the optimal number of clusters except
when the FCM algorithm is applied. Only, PC cor-
rectly identifies the optimum number for both cluster-
ing algorithms: FCM and FCS.
After clustering the five datasets using the FCM
and FCS, we compare them in terms of the PC and
CE, FS, XB and BS values. Results show that PC
yields the optimal number of clusters three times, CE
identifies the correct number of clusters three times
and only with FCM. Since, FS yields the correct num-
ber of clusters only one time and XB does not yield
the correct number of clusters for any dataset, they
are the most unreliable indices. BS yields the optimal
number of clusters four times. Hence, we confirm re-
sults of (Cho and Yoo, 2006) that the Bayesian score
is the most reliable clustering validity measure. How-
ever, none of the above mentioned indices correctly
finds the optimal number of clusters for all data sets.
Therefore, a suitable index must be selected for each
data.
4 CONCLUSIONS
In order to evaluate fuzzy partitions of two clustering
algorithms, FCM and FCS, four conventional valid-
ity measures and Bayesian validation are used on five
datasets. Results show the good performance of the
Bayesian score as a cluster validity index and demon-
strates that in comparison with conventional fuzzy
indices the Bayesian validation leads to superior re-
sults. We conclude that none of the above mentioned
indices leads to the correct number of clusters for
all mentioned datasets. Hence, fuzzy clustering re-
quires more investigations, where most clustering al-
gorithms may not provide satisfactory result because
no single validity measure works efficiently on differ-
ent kinds of datasets. As future research, fuzzy clus-
tering analysis from a multiple objective optimization
perspective where the search should be performed
overa number of often conflicting objectivefunctions,
needs to be studied.
REFERENCES
Bezdek, J. (1974). Cluster validity with fuzzy sets. Journal
of Cybernetics and Systems, 3(3):58–72.
Carvalho, F. (2006). A fuzzy clustering algorithm for
symbolic interval data based on a single adaptive eu-
clidean distance. In ICONIP (3), pages 1012–1021.
Cho, S. and Yoo, S. (2006). Fuzzy bayesian validation
for cluster analysis of yeast cell-cycle data. Pattern
Recognition, 39(12):2405–2414.
CONVENTIONAL AND BAYESIAN VALIDATION FOR FUZZY CLUSTERING ANALYSIS
139

Dave, R. (1990). Fuzzy shell-clustering and applications to
circle detection in digital image. International Journal
of General Systems, 16(12):343–355.
Fukuyama, Y. and Suengo, M. (1989). A new method of
choosing the number of clusters for thefuzzy c-means.
Proceedings of Fifth Fuzzy Systems Symposium, pages
247–250.
Graves, D. and Pedrycz, W. (2010). Kernel-based fuzzy
clustering and fuzzy clustering: A comparative exper-
imental study. Fuzzy Sets and Systems, 161(4):522–
543.
Halkidi, M., Batistakis, Y., and M.Vazirgiannis (2001). On
clustering validation techniques. J. Intell. Inf. Syst.,
17(2-3):107–145.
Klawonn, F. and Hoppne, F. (2009). Fuzzy cluster analy-
sis from the viewpoint of robust statistics. Studies in
Fuzziness and Soft Computing, 243(19):439–455.
Wang, W. and Zhang, Y. (2007). On fuzzy cluster validity
indices. Fuzzy Sets and Systems, 158(19):2095–2117.
Xie, X. and Beni, G. (1991). A validity measure for fuzzy
clustering. IEEE Trans. Pattern Anal. Mach. Intell.,
13(8):841–847.
Yang, X., Cao, A., and Song, Q. (2006). A new cluster
validity for data clustering. Neural Processing Letters,
23(3):325–344.
ICFC 2010 - International Conference on Fuzzy Computation
140
