
GENETIC EVOLUTION OF ‘SORTING’ PROGRAMS THROUGH
A NOVEL GENOTYPE-PHENOTYPE MAPPING
Daniela Xhemali, Christopher J. Hinde and Roger G. Stone
Department of Computer Science, Loughborough University, Loughborough, U.K.
Keywords: Genetic programming, Genotype-phenotype mapping, XML, Regular expressions, Software programs.
Abstract: This paper presents an adaptable genetic evolutionary system, which includes an innovative approach to
mapping genotypes to phenotypes through XML rules. The evolutionary system was originally created to
evolve Regular Expressions (REs) to automate the extraction of web information. However, the system has
been adapted to work with a completely different domain – Complete Software Programs – to demonstrate
the flexibility of this approach. Specifically, the paper concentrates on the evolution of ‘Sorting’ programs.
Experiments show that our evolutionary system is successful and can be adapted to work for challenging
domains with minimum effort.
1 INTRODUCTION
Genetic Programming (GP) can be defined as “a
systematic, domain-independent method for getting
computers to automatically solve problems starting
from a high-level statement of what needs to be
done” (Langdon et al., 2008). GP research has
attracted attention in various fields such as: game
strategies (Keaveney & O’Riordan, 2009), military
defence (Jackson, 2005), plant biology (Dyer &
Bentley, 2002), electronics (O’Neill et al., 2001),
railway platform allocation (Clarke et al., 2009),
spam filtering (Conrad, 2007), feature extraction
from media files (Hsu, 2007; Klank et al., 2008) etc.
An area that has also managed to secure the
attention of GP is the automation of Web
Information Extraction (WIE) (Atkinson-Abutridy et
al., 2004; Barrero et al., 2009; Xhemali et al., 2010-
b). The research presented in this paper was
originally set up to evolve Regular Expressions
(REs) to automate the extraction of web information,
specifically training course information such as:
course names, dates, locations and prices. The
details of this part of the research, including
experimental results, were covered previously
(Xhemali et al., 2010-b), thus they will not be
covered again in this paper.
This paper focuses on a specific part of GP – the
genotype to phenotype mapping. Previous work
(Xhemali et al., 2010-a) gave details of our
innovative approach in relation to its application to
REs and to Complete Software Statements (Withall
et al., 2008) such as FOR loops, IF statements etc.
This paper concentrates on the genotype to
phenotype mapping process in relation to Complete
Software Programs. Specifically, experiments are
carried out to test the complete GP process for the
evolution of ‘Sorting’ programs. ‘Sorting’ programs
were chosen in order to demonstrate the flexibility
of this approach, as such programs represent an
entirely different domain to REs and a challenging
problem for GP systems.
2 RELATED RESEARCH
The genotype-phenotype mapping relates to the way
individuals in a population are represented, as this
can have a significant effect on the performance of
GP. A genotype represents each individual in the
search space, whereas its phenotype represents the
individual in the solution space (Banzhaf, 1994).
Some research, particularly earlier GP research, does
not make a distinction between genotypes and
phenotypes (Koza, 1992; Whigham, 1995; Conrad,
2007, Snajder et al., 2008 etc.). Individuals in each
genetic population remain the same throughout the
evolution process. In these works the search space
and the solution space are identical.
In 1994, Banzhaf suggested the separation of the
two spaces and introduced his work on the
genotype-phenotype mapping. The separation
190
Xhemali D., Hinde C. and Stone R..
GENETIC EVOLUTION OF ‘SORTING’ PROGRAMS THROUGH A NOVEL GENOTYPE-PHENOTYPE MAPPING .
DOI: 10.5220/0003078401900198
In Proceedings of the International Conference on Evolutionary Computation (ICEC-2010), pages 190-198
ISBN: 978-989-8425-31-7
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

involves the encoding of the individuals to a form
known as the genotype, which is later on decoded
back to the corresponding program, referred to as the
phenotype. Since then, many other researchers have
embraced the separation into genotypes and
phenotypes (Keller & Banzhaf, 1996; Withall et al.,
2008; Clarke et al., 2009 etc.). This separation
simplifies and increases the efficiency of certain
genetic operations such as: reproduction and
mutation, because these would no longer be
constrained by the parameters used in the program
being evolved. In genotype-phenotype based GP,
genetic operators such as Crossover and Mutation
would be performed on the genotype, whereas other
processes, such as the Fitness scoring, would be
performed on the phenotype. Sections 3.2 and 3.3
explain this concept further through examples.
On the downside, however, this adds an
additional step to the genetic evolution process – the
translation or mapping of the genotypes to their
corresponding phenotypes. This step occurs after the
genetic reproduction stage (i.e. the crossover and
mutation) and before the Fitness test can take place.
There are researchers who criticise the separation
into genotypes and phenotypes (Moore, 2000). The
main concern expressed is that the conversion
process of a mutated genotype into the phenotype
may result in anomalies that could potentially lead to
invalid solutions. A direct mapping between the
encoded program (genotype) and the actual program
(phenotype) is therefore vital to ensure the validity
of the solutions (Rothlauf, 2006; Withall et al.,
2008).
The following discusses different methods that
have been used to achieve the mapping process.
Banzhaf (1994; 2006) represented his genotypes
as linear binary strings. The mapping stage then
processed these genotypes from left to right in 5-bit
sections, where each 5-bit code mapped to a pre-
specified symbol. For example: 00000 mapped to
PLUS, 00100 mapped to POW, 11000 mapped to
variable X etc. The first bit indicated whether the
code represented a function (PLUS, POW etc.) or a
terminal (X, Y etc.). The research also discussed
their concern about generating constant numbers.
Koza (1992) had solved this problem by defining
“random ephemeral constants” where constants are
only generated once for a particular program and
then reused wherever they are needed within that
program.
Keller (1996) continued in the footsteps of
Banzhaf, concentrating on providing experimental
evidence for choosing the genotype-phenotype
approach instead of the normal GP approach. Keller
used the LALR (Look Ahead LR) parser for the
repairing stage of the genotype-phenotype mapping
process. LALR parsers scan the input from left to
right and construct a rightmost derivation in reverse
(Aho and Ullman, 1979).
There was a certain amount of redundancy in the
genetic coding in both Banzhaf’s and Keller’s
works. They both admitted that, in their works,
different binary strings could correspond to the same
symbol, which could lead to inconsistencies e.g. 000
and 100 both mapping to ‘a’.
A slightly different genotype representation is
seen in the work of Withall et al. (2008). In here
genotypes are represented as linear blocks of
integers. Each block consists of exactly four
integers, each integer representing a different gene.
Although both research works used fixed-length
genomes, in the work of Banzhaf (1994; 2006) the
resulting phenotypes could vary in length, whereas
in (Withall et al., 2008) they remained fixed.
However, Withall allowed for variable-length
genomes through padding, whereby shorter program
structures or statements ignored outstanding genes.
The first integer in Withall’s genotype always
determines the type of function that follows.
Grosan and Abraham (2009) worked with multi-
chromosome genotypes. The number of
chromosomes was varied. However, the number of
genes per chromosome was fixed. In this research,
each gene corresponded to either a terminal: T = {a,
b, c, d} or a function: F = {+, *}. A function gene
also included pointers towards the function
parameters to tell the system which terminals were
to be manipulated by the function. Also, the first
gene of the chromosome was always a terminal. This
was to ensure that only syntactically correct
programs are evolved. Very differently from above,
Yosif et al. (2010) introduced the novel approach of
adapting a support vector machine to predict
phenotypes from genotype data.
Similarly to Withall et al. (2008), the genotype in
our research is represented as a string of integers.
There are no fixed length genomes determined
however; instead the genotype can contain any
number of genes. The direct mapping of these
integers to the corresponding structures is a
chieved
through an innovative approach involving XML
rules. The first gene in the string determines the
XML rule to be followed, which in itself guides the
mapping of the rest of the genes into a valid
phenotype. This is explained in detail in section 3.2.
GENETIC EVOLUTION OF 'SORTING' PROGRAMS THROUGH A NOVEL GENOTYPE-PHENOTYPE MAPPING
191

3 GENOTYPES TO
PHENOTYPES
As previously mentioned, some details about this
research, including the GP representation chosen and
a thorough explanation of our novel genotype-
phenotype mapping approach used on the REs
domain, was published in a previous paper (Xhemali
et al., 2010-a), thus they will not be repeated here.
The rest of this paper concentrates on the application
of the genotype-phenotype approach to a completely
different domain – that of Software Programs,
specifically ‘Sorting’ programs – in order to
illustrate the flexibility of our approach.
Additionally, the fitness function for the ‘Sorting’
programs is presented and experimental results are
discussed.
The examples in this paper are based on the work
of Withall et al. (2008). They are kept as close to the
original as possible in order to ensure their integrity.
One main difference however, is that in Whithall’s
work the evolved programs were in Perl, whereas in
this paper their validity is ensured against VB script.
Although the evolutionary system is developed in
VB.NET with a MS SQL Server database backend,
VB Script was used in order to extend the
application to execute each evolved program, as
these are obviously not compiled into the
application. The .NET framework allows for the
execution of dynamic code through the
System.CodeDom.Compiler and the appropriate
namespaces, however, this is more complicated and
slower during execution than utilising the Microsoft
Script Control from VB.NET.
3.1 ‘Sorting’ Program
The ‘Sorting’ program was chosen, because it is a
popular, well known program and a standard
Computer Science problem due to its higher
complexity over other small software programs such
as: ‘Sum finder’, ‘Maximum value finder’ etc.
The aim of a ‘Sorting’ program is to order a list
of integers or characters in ascending order. The
output of a ‘Sorting’ program is therefore another
list, rather than a single value.
The ‘Sorting’ programs evolved in this research
are concerned with the sorting of lists of integers.
Figure 1 shows a specification of the ‘Sorting’
problem.
The following explains the general details behind
the genotype-phenotype mapping for software
structures, as well as the additional statements and
genes needed for the ‘Sorting’ program.
Figure 1: Specification of ‘Sort’ (Withall et al., 2008).
3.2 XML Mapping
Our genotype-phenotype mapping consists of two
main components: the XML rules, which guide the
system through the mapping process and the
Repairing function, which makes sure that the
evolved programs are syntactically correct. The
genotype-phenotype mapping process is as follows:
Pseudo-code: Genotype-Phenotype Mapping
1) Determine the XML rule to follow
2) Follow the chosen XML rule to the end
3) IF the Genotype has fewer genes than the rule
requires
a) Follow the rule for the number of genes
available
b) Repair outcome to create a valid partial
solution.
4) IF the Genotype has enough genes for the XML
rule
a) Follow all the components in the rule
b) Repair outcome (if necessary) to create a
valid and complete solution.
5) IF the Genotype has more genes than the rule
requires
a) Follow the same steps as above (4a and 4b)
b) Ignore the rest of the genes in the Genotype
Note that this is not a character by character
evolution, because this would increase the search
space and dramatically increase the execution time.
Instead, programs are divided into two collections:
Variables (e.g. “tmp1”, “tmp2”, “tmp3”) and
Comparisons (e.g. “>”, “<”, “!=” etc.). Each evolved
gene is translated to an element of one of these
collections. There is a separate XML rule for each
software structure (e.g. “FOR”, “IF ... THEN ...
ELSE”, “ADD”, “ASSIGN” etc.). Each rule is
ICEC 2010 - International Conference on Evolutionary Computation
192
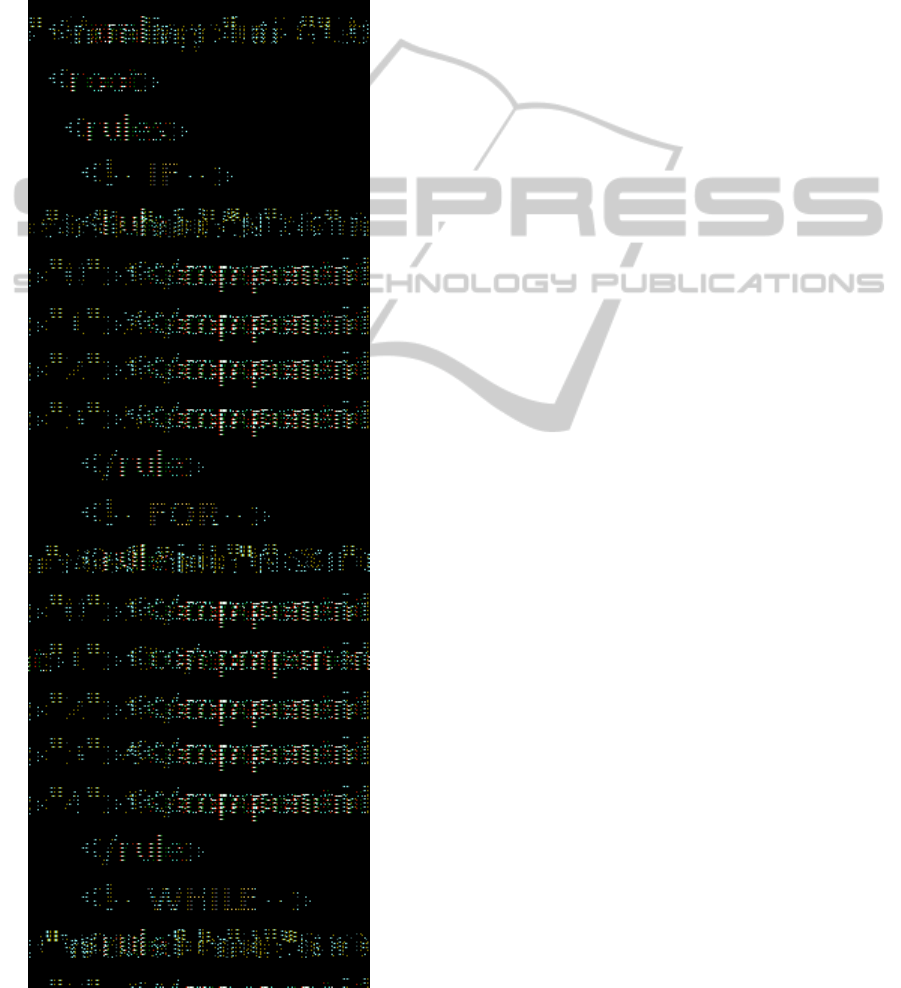
composed of a number of components, which guide
the system through the translation of each gene to
the corresponding software structure (Figure 2). For
example, the IF ... THEN structure, of format (IF
variable
1
comparison variable
2
THEN), requires
three evolvable genes: two variables and one
comparison. Table 1, Table 2 and Figure 3 illustrate
this scenario by giving an example of the genotype-
phenotype mapping process.
Figure 2: Sample of XML Rules.
Each component refers to either the elements in
the above two collections, or to other predefined
elements that do not need to be evolved and as such
do not require the use of any extra genes, such as:
the different operators associated with each
programming structure (e.g. “+” is always associated
with “Add”; “=” is always associated with “Assign”,
thus these do not need to be evolved) or the
keywords required by the programming language
chosen – in this case VBScript – in order to create
syntactically correct code (e.g. “THEN”, “TO” etc).
The Repairing function is responsible for making
sure that all the different software structures are
combined correctly to create a syntactically correct
and complete software program. Figure 2 shows a
sample of the XML rules and components needed to
guide the genotype-phenotype stage of the genetic
evolution of software programs. Table 1 shows a
sample Genotype to be translated using the
information in Figure 2.
Note that the first gene in the genotype is
always associated with the XML rule choice. The
modulo function is used for this reason. In the above
example (Table 1), the value of the first gene is 10.
This represents the software structure to be used. In
this case, there are five different rules in the XML
file, so 10 mod 5 = 0 means that the structure chosen
is an ‘IF’. This structure contains four different
components (Figure 2). The first three components
require the use of a gene to choose from either the
‘variables’ or the ‘comparisons’ collections. The
‘variables’ collection has three elements, whereas
the ‘comparisons’ has four, therefore, 27 mod 3 = 0,
7 mod 4 = 3 and 13 mod 3 = 1 give elements ‘x’, ‘<’
and ‘y’ respectively.
The fourth component (id=“5”) tells the
Repairing function that the THEN keyword is
required next. The ‘THEN’ keyword is a mandatory
requirement for IF statements in VB.NET or
VBScript, thus this component does not need to be
evolved and as such does not require the use of an
extra gene from the genotype.
The nested = “true” attribute seen in Figure 2
indicates that the IF structure, differently from one-
line statements, such as ‘Add’ or ‘Multiply’, expects
other statement(s) inside. The following gene (gene
19) in the genotype is therefore used to determine
the statement type to be nested in this IF. Therefore,
19 mod 5 = 4 means that the next statement is ‘Add’.
The remaining components are dealt with in the
same manner (see Table 2).
Once all the required genes have been decoded,
the resulting phenotype is repaired to ensure it is
syntactically correct. The Repairing function is an
GENETIC EVOLUTION OF 'SORTING' PROGRAMS THROUGH A NOVEL GENOTYPE-PHENOTYPE MAPPING
193

independent function, which scrutinises the
phenotype created in order to guarantee the syntactic
validity of the solution. This function is in charge of
tasks like: closing software structures appropriately
(e.g. FOR loops in VB.NET need to end in NEXT
for them to be valid); adding the necessary
operators, which do not need to be evolved (e.g. the
‘Add’ (“+”) and the ‘Assign’ (“=”) operators);
tidying up the phenotype in cases when there are
fewer genes available than required by the XML
rule; adding header and footer information about a
solution such as: variable declaration or variable
return etc. All this is achieved through the use of a
STACK programming structure, which works in a
LIFO (Last In First Out) manner. Figure 3 shows the
complete software structure (phenotype) for the
above example. The additional symbols and
programming keywords added by the repairing
function are shown encircled.
Table 1: Genotype.
10 27 7 13 19 9 63 4
Table 2: Genotype to Software Statement Mapping.
Comp.
No.
Component
Gene
Used
Modulo Translation
- - 10 0 If
1 Variable 27 0 x
2 Comparison 7 3 <
1 Variable 13 1 y
5
Then-
Keyword
- - Then
- - 19 4 Add
1 Variable 9 0 x
11 Assign - - =
1 Variable 63 0 x
7 Addition - - +
1 Variable 4 1 y
If x < y Then
x = x + y
End If
Figure 3: Phenotype.
Updating the XML rules, once written, would
require little effort, because software statements and
structures are rigid in the number of components
needed and the order in which they are needed. For
example, the Add statement mentioned above may
sum up more than two variables, however, there will
always be need for one variable to which this sum is
assigned, one ‘Assign’ operator and one or more
‘Addition’ operators. Adding new XML rules for
new statements or structures would be equally as
effortless, because it would only require the addition
of the different components for that structure to the
XML rules as well as any additional variables or
comparisons that may be required.
The changes that were made to our Genotype-
Phenotype mapping process for the ‘Sorting’
software programs involved the simple addition of
one more structure and a few more variables and
comparison values. Specifically, a special nested
FOR loop was added to the current software
structures, similarly to Withall et al. (2008), which
includes two nested FOR loops in the following
format:
FOR (var
1
= 0 TO var
2
)
FOR (var
3
=
var
1
+1 TO var
2
)
...
The above could have been achieved through the
existing FOR loop structure (Figure 2), however, we
chose to add the nested FOR loop, because this is a
standard structure used when comparing elements in
a list and it results in faster execution of the
evolution process.
Due to the ‘Sorting’ program dealing with lists or
arrays of integers, a few more variable types needed
to be added to distinguish between normal variables
and array variables (since a normal variable x is
entirely different from the variable array(x)). This is
to maintain the accuracy of the evolved solutions.
The Experimental Results section presents and
discusses the results of our experiment.
3.3 Fitness Function
Similarly to Withall et al. (2008), the fitness
function in this research has been simplified to only
compare adjacent items in the list of integers rather
than all the possible pairs in the list. This decision
was made in order to speed up the evolution process.
The code for the ‘Sorting’ program is presented
above to show its simplicity. Note that the necessary
header information such as variable declaration and
initialisation are left out for clarity. Also note that
this function is only called if the post-evolution list
of integers contains the same elements as the pre-
evolution list of integers.
ICEC 2010 - International Conference on Evolutionary Computation
194

Code: Fitness Function for ‘Sorting’
If list.Length > 0 Then
For i As Int32=0 To list.Length -
2
If list(i) <= list(i + 1) Then
fitness += 1
End If
Next
End If
If list.length > 1 Then
fitness = fitness/(list.Length-1)
Else ‘The list has only got one
integer
fitness = 1
End If
4 EXPERMIMENTAL RESULTS
Details about the evolutionary system used in this
research were discussed in a previous paper
(Xhemali et al., 2010-b) however, the main
parameters used by the system are summarised in
here to help the reader fully appreciate the
experimental results presented:
Figure 4: Complete ‘Sorting’ Solution: (1).
Population size: 10
Tournament size: 40%
Fitness Target: 1
Uniform Crossover Rate: 50%
Mutation Rate: 1 gene per genotype
Initial Population: Random
The following gives the results from the
experiments set up for the evolution of ‘Sorting’
programs. In order to maintain the similarity with
the work of Withall et al. (2008), there were ten runs
of the experiment, each with a maximum number of
generations of 50,000. None of the experiments
needed this many generations however, and each run
produced a valid ‘Sorting’ program.
Figure 4 shows one of the ‘Sorting’ programs
produced by the system. Note that this example
shows the full VBScript code executed from within
VB.NET 2008. The header and footer of the solution
(shown in Figure 4 within the dotted rectangle) was
added to the solution by the Repairing function. The
part of the code shown within the solid rectangle
was evolved by the system (and tidied up by the
Repairing function, as described in section 3.2).
All experiments presented in this paper were
based on the sorting process of a fixed list of seven
integers – a(1, 3, 2, 4, 8, 5, 9) – however, further
experiments were carried out with different integer
lists to ensure that the results in this paper were not
somehow influenced by the integers chosen.
Table 3 gives information about five of the
‘Sorting’ solutions evolved (Fitness Score: 1), which
were also generated by Withall et al. (2008). The
information includes the number of generations
(Gens) taken for the solution to be generated and the
amount of time taken to arrive to this solution. The
remaining five solutions (6-10), also with a Fitness
Score of 1, refer to additional ‘Sorting’ solutions
evolved during the experiments for this paper.
Table 3: ‘Sorting’ Results.
Solution
No.
Gens
Gens
(Withall)
Time
Time
(Withall)
1 35467 47975 57’33” 1h04’28”
2 16200 14189 25’25” 19’22”
3 8950 8219 12’08” 10’22”
4 15982 16312 23’12” 21’57”
5 762 5834 2’36” 8’00”
6 20050 - 27’31” -
7 739 - 2’19” -
8 1690 - 4’48” -
9 559 - 1’45” -
10 93 - 0’15” -
GENETIC EVOLUTION OF 'SORTING' PROGRAMS THROUGH A NOVEL GENOTYPE-PHENOTYPE MAPPING
195

The solutions themselves (without the header or
footer information), including the five that were
different from the solutions generated by Withall et
al. (2008) are listed in Appendix A.
In relation to the results in Table 3, it is
important to note that comparing the results of
different evolutionary systems is not straightforward
and by no means definitive. Despite experiments
being carried out on the same domain, a touch of
luck is also involved in getting to a perfect solution
quickly from evolutionary experiments. This is
because, depending on the crossover of the genes
and particularly on the (random) gene that gets
chosen for mutation and the outcome of the mutation
itself, a perfect solution may not be reached at the
same time by different systems. Furthermore, even
experiments carried out on the same system at
different times, may not arrive at the same solution
at the exact same generation. Keeping this in mind,
Table 3 shows that Withall et al. (2008) need fewer
generations than this research for two of the above
solutions (2 and 3). This research needs fewer
generations for solutions 1, 4 and 5.
One thing is evident however, that Withall’s
timings (time needed per generation) are lower than
those in this research. As previously mentioned
Withall’s genetic evolutionary system was all
written in Perl, thus there was no need for
converting the evolved code to a Scripting language
first to achieve dynamic execution, since Perl is
already a scripting language. The system in this
research however, was written in VB.NET and
includes the additional step of forcing the execution
of the solution as VBScript from within the VB.NET
application. This affects the overall execution speed.
Furthermore, it was observed during the experiment
that VBScript displayed a message box to the user
each time the script took longer than normal to
execute. The user was then asked to choose whether
to allow the script to continue running or end it and
allow the system to move onto the next script.
We managed to change the VBScript control to
make the decision by itself, without involving the
user. Although this has sped up the execution, it is
still an extra step that VBScript has to do behind the
scenes, which increases the overall execution time
for the evolutionary system.
A potential solution may be to change the system
to execute the dynamic programs in Perl instead of
VBScript and see if this makes a difference to the
above timing issue. Another solution may be to
allow .NET itself to execute the dynamic code
through the inbuilt System.CodeDom.Compiler,
however, this is a more complicated solution, which
may still result in time wastage due to the additional
manipulation that VB.NET will have to make to
itself to compile the dynamic code at runtime.
4 CONCLUSIONS
This paper has discussed an innovative approach to
mapping genotypes to phenotypes through XML
rules in relation to software statements and
structures as well as complete software programs.
Utilising XML gives this technique many
advantages including: improved readability,
compatibility with many programming languages,
portability and extendibility (XML is not restricted
to a limited set of keywords defined by the
proprietary vendors, which aids the process of
creating rules of different levels of complexity).
Experiments were set up to test the complete
evolutionary system on the evolution of ‘Sorting’
programs. The ‘Sorting’ program was chosen
because it represents a well known, standard
Computer Science problem, which is complex
enough to really test the evolutionary system.
Our evolutionary system was originally created
to deal with the evolution of Regular Expressions.
However, it was discovered, that it was easily
adaptable to other domains, including that of
Complete Software Structures. In fact, the only two
areas that needed to be changed to make the system
work for the new domain, were the XML rules and
the Fitness function.
The experimental results from the evolution of
‘Sorting’ programs highlight the efficacy and
flexibility of our system, despite the complexity of
the ‘Sorting’ problem for GP systems.
However, further testing needs to be done to
ensure the reliability of this approach for other
complex programs.
ACKNOWLEDGEMENTS
We would like to thank the team at ATM for their
support. Also, thank you to ATM, CICE and
Loughborough University for funding our work. We
would also like to thank Daniel Sills for his help
with some technical .NET concepts.
REFERENCES
Aho, A. V. & Ullman, J. D. 1979. Principles of Compiler
ICEC 2010 - International Conference on Evolutionary Computation
196

Design. Addison Wesley, ISBN 0-201-00022-9.
Atkinson-Abutridy, J., Mellish, C., Aitken, S. 2004.
Combining information extraction with genetic
algorithms for text mining. IEEE Intelligent Systems.
19(3), pp. 22-30.
Banzhaf, W. 1994. Genotype-Phenotype-Mapping and
Neutral Variation – A case study in Genetic
Programming. Proceedings of the International
Conference on Evolutionary Computation. Springer-
Verlag, pp.322-332.
Banzhaf, W. 2006. genotype-phenotype-mapping and
neutral variation – A case study in Genetic
Programming. Lecture Notes in Computer Science,
Springer Berlin, 866, pp. 322-332.
Barrero, D., Camacho, D. & R-Moreno, M. 2009.
Automatic Web Data Extraction Based on Genetic
Algorithms and Regular Expressions. Data Mining
and Multi-agent Integration. Springer-Verlag, pp. 143.
Clarke, M., Hinde, C. J., Withall, M. S., Jackson, T. W.,
Philips, I. W., Brown, S. & Watson, R. 2009.
Allocating Railway Platforms using a Genetic
Algorithm. Research and Development in Intelligent
Systems XXVI. Springer London, pp. 421-434.
Conrad, E. 2007. Detecting Spam with Genetic Regular
Expressions. SANS Institute Reading Room. Available:
http://www.giac.org/certified_professionals/practicals/
GCIA/00793.php.
Dyer, J. & Bentley, P. 2002. PLANTWORLD: Population
Dynamics in Contrasting Environments. In Cantu-Paz
E., GECCO, pp. 122-129.
Grosan, C. & Abraham, A. 2008. Evolving Computer
Programs for Knowledge Discovery. Social Science
Research Network (SSRN).
Hsu, P-H. 2007. Feature extraction of hyperspectral
images using wavelet and matching pursuit. ISPRS
Journal of Photogrammetry and Remote Sensing.
Elsevier Science, Amsterdam, 62 (2), pp. 78-92.
Jackson, D. 2005. Evolving Defence Strategies by Genetic
Programming. In Lecture Notes in Computer Science.
Springer Berlin, 3447, 281-290.
Keaveney, D. & O’Riordan, C. 2009. Evolving Robust
Strategies for an Abstract Real-time Strategy Game.
Proceedings of the 5
th
International Conference on
Computational Intelligence and Games. pp. 371-378.
Keller, R. E. & Banzhaf, W. 1996. Genetic Programming
using Genotype-Phenotype Mapping from Linear
Genomes into Linear Phenotypes. Proceedings of the
First Annual Conference on Genetic Programming,
California. pp. 116-122.
Klank, U., Padoy, N., Feussner, H. and Navab, N. 2008.
Automatic feature generation in endoscopic images.
International Journal of Computer Assisted Radiology
and Surgery. Springer, 3, pp. 331-339.
Koza, J. R. 1992. Genetic Programming: On the
Programming of Computers by Means of Natural
Selection. MIT Press.
Langdon, W., Poli, R., McPhee, N. & Koza, J. R. 2008.
Genetic Programming: An Introduction and Tutorial
with a Survey of Techniques and Applications. In
Studies in Computational Intelligence. Springer,
Berlin, 115, pp. 927-1028.
Moore, J. P. 2000. Exploring and Exploiting Models of the
Fitness Landscape: A Case against Evolutionary
Optimization. PhD Thesis, University of Plymouth.
O’Neill, M., Brabazon T., Ryan, C. & Collins J. J. 2001.
Developing a Market Timing System using
Grammatical Evolution. Proceedings of GECCO.
Rothlauf, F. 2006. Representations for Genetic and
Evolutionary Algorithms. Springer-Verlag New York.
Snajder, J., Basic, B. D., Petrovic, S. & Sikiric, I. 2008.
Evolving new lexical association measures using
genetic programming. Proceedings of the Association
for Computational Linguistics. Ohio, pp. 181-184.
Whigham, P. A. 1995. Grammatically-based Genetic
Programming. Workshop on Genetic Programming.
Withall, M. S., Hinde, C. J. & Stone, R. G. 2008. An
improved representation for evolving programs.
Journal of Genetic Programming and Evolvable
Machines. Springer Netherlands, 10(1), pp. 37-70.
Xhemali, D., Hinde, C. J. & Stone, R. G. 2009-a. Domain-
Independent Genotype to Phenotype Mapping through
XML Rules. International Journal of Computer
Science Issues, 7(3).
Xhemali, D., Hinde, C. J. & Stone, R. G. 2010-b. Genetic
Evolution of Regular Expressions for the Automated
Extraction of Course Names from the Web.
Proceedings of the International Conference on
Genetic and Evolutionary Methods. Las Vegas.
Yosif, N., Gramm, J., Wang, Q., Noble, W., Karp, R. &
Sharan, R. 2010. Prediction of Phenotype Information
from Genotype Data. Communications in Information
and systems. 10(2), pp. 99-114.
APPENDIX A
These are the ‘Sorting’ programs generated in this
research. Solution (1) was shown in
Figure 4.
Solutions (1-5) match those obtained by Withall et
al. (2008), whereas the remaining (6-10) are new.
Note that parts of the following programs may
look different to ‘hand written’ code for ‘Sorting’
programs. Solutions (3) and (9) are the closest to the
conventional ‘hand coded’ version.
‘Sorting’ Solution: (2)
GENETIC EVOLUTION OF 'SORTING' PROGRAMS THROUGH A NOVEL GENOTYPE-PHENOTYPE MAPPING
197

‘Sorting’ Solution: (3)
‘Sorting’ Solution: (4)
‘Sorting’ Solution: (5)
‘Sorting’ Solution: (6)
‘Sorting’ Solution: (7)
‘Sorting’ Solution: (8)
‘Sorting’ Solution: (9)
‘Sorting’ Solution: (10)
ICEC 2010 - International Conference on Evolutionary Computation
198
