
SUPPORTING SCIENTIFIC BIOLOGICAL APPLICATIONS
WITH SEAMLESS DATABASE ACCESS
IN INTEROPERABLE E-SCIENCE INFRASTRUCTURES
Sonja Holl, Morris Riedel, Bastian Demuth, Mathilde Romberg and Achim Streit
Juelich Supercomputing Center, Forschungszentrum Juelich, 52428 Juelich, Germany
Keywords:
Bioinformatics, Database access, e-Science, Life science.
Abstract:
In the last decade, computational biological applications have become very well integrated into e-Science
infrastructures. These distributed resources, containing computing and data sources, provide a reasonable en-
vironment for computing and data demanding applications. The access to e-Science infrastructures is mostly
enstablished via Grids, where Grid clients support scientists using different types of resources. This paper
extends an instance of the infrastructure interoperability reference model to remove the lack by adding cen-
tralized access to distributed computational and database resources via a graphical Grid client.
1 INTRODUCTION
Computational biological applications have evolved
as a very important element in bioinformatics that
bring insights in biological phenomena. However,
they are typically very demanding in terms of com-
putational capabilities, since they use complex math-
ematical models and simulations, and in terms of data
capacities, since they require and produce a lot of
data. In the context of computational capabilities
they use high-performance computing (HPC), high-
throughput computing (HTC) and in the context of
data capacities, biological applications use a wide
variety of data sources. In more detail, researchers
need to extract information from large collections
of data to analyse experiment outputs as well as to
share and reuse data results from different geograph-
ical locations. That raises the demand of a seam-
less and scalable access to different distributed re-
sources, especially computing and data resources. In
order to tackle these requirements, enhanced Science
(e-Science) (Taylor et al., 2006) infrastructures have
become successfully established as an interoperable
distributed environment for computational biological
applications (Baldridge and Bourne, 2003).
Today, e-Science infrastructures have been realized
by ’next generation infrastructures’, and represented
by Grids today (Berman et al., 2003). Grids provide
seamless and secure collaborations over wide-area
networks in key areas of science and can be accessed
via middleware systems. Examples are the middle-
ware UNICORE (Streit et al., 2009), ARC (Ellert
et al., 2007), Globus Toolkit (Foster, 2005), or
gLite (gLite, 2009). Scientists, using e-Science en-
vironments are normally supported by feature rich
graphical clients. But the majority of these clients
lack the support of centralized access to biological
data and distributed computing resources that are both
required in bioinformatics domains.
In this paper, we describe the integration and
support of biological databases with a focus on
how this can be realized in the existing model of
the UNICORE middleware and the UNICORE Rich
Client (URC, 2009) (URC). This extensions enable
biologists within the URC to use effectively interop-
erable infrastructure setups that are based on the pro-
posed infrastructure interoperability reference model
(IIRM) (Riedel et al., 2009).
The remainder of this paper is structured as fol-
lows. In Section 2, we introduce the infrastructure in-
teroperability reference model, which acts as the ref-
erence model for our infrastructure setups. The de-
sign of the database support in a graphical client is
presented in Section 3. In Section 4 we present a use
case application. Finally, we compare related work in
Section 5 and conclude the paper in Section 6.
203
Holl S., Riedel M., Demuth B., Romberg M. and Streit A. (2010).
SUPPORTING SCIENTIFIC BIOLOGICAL APPLICATIONS WITH SEAMLESS DATABASE ACCESS IN INTEROPERABLE E-SCIENCE INFRASTRUC-
TURES.
In Proceedings of the First International Conference on Bioinformatics, pages 203-206
DOI: 10.5220/0002695002030206
Copyright
c
SciTePress

2 THE INFRASTRUCTURE
INTEROPERABILITY
REFERENCE MODEL
Over the years, various types of production e-Science
infrastructures evolved that can be classified into dif-
ferent categories. While there are infrastructures
driven by HPC such as TeraGrid or DEISA (DEISA,
2009), there are also infrastructures that rather sup-
port HTC such as EGEE (Laure and Jones, 2008).
In addition hybrid infrastructures such as D-Grid (D-
Grid, 2009) exit, but more notably a significant prob-
lem is that nearly all of these different infrastructures
deployed non-interoperable Grid middleware systems
in the past.
HTC-Driven
Applications
EGEE Resources
Production
e-infrastructures
Different
computing
paradigms
using joint
data storages
DEISA Resources
HPC-Driven
Applications
Scientific
Data
Implemented
refined & tuned
standards in
Grid technologies
& missing links
Infrastructure Interoperability Reference Model Set of Standards
Numerous
clients
for transparent
infrastructure
access
Scientific-area specific clients and portals
Tuned
OGSA-BES
Security
GLUE2
GridFTP
GridSphere Scientific Clients
…
UNICORE Rich Client
Joint Data Storages
Tuned
SRM
Tuned
WS-DAIS
Figure 1: The interoperability infrastructure reference
model (IIRM) that realizes the interoperability of the e-
science infrastructure EGEE and DEISA.
Based on the experiences and lessons learned from
numerous international interoperability efforts, we
have defined an interoperability infrastructure refer-
ence model (IIRM) (Riedel et al., 2009), which is
based on well-known open standards.
The added value of the IIRM is that it defines miss-
ing links, refinements, and improvements of these
standards gained from interoperability experiences
with real applications. As shown in Figure 1, this
paper focuses on one IIRM instance that realizes the
interoperability of the e-Science infrastructure EGEE
and DEISA as described in (Riedel et al., 2008), but
highlights its potential to conduct e-Science in the
field of life sciences leveraging the seamless access
to HTC and HPC resources with a particular focus on
the database aspects of it. Apart from other scientific
fields, the life science community especially benefits
from joint data storages to store and re-use interme-
diate data during different computation phases that
often use different computing paradigms (i.e. HTC,
HPC).
While different clients can access the IIRM-based
Grid technologies (e.g. UNICORE, ARC, gLite) as
shown in Figure 1, this paper describes one particu-
lar usage model with the UNICORE Rich Client in
the context of database and computational resource
access.
3 SUPPORTING DATABASE
ACCESS
Since biological applications are widely used in
e-Science environments, the demand for easy accessi-
ble distributed biological data in conjunction with dis-
tributed computational resources in e-Science infras-
tructures is steadily increasing. This demand is based
on the fact that many highly scalable, mostly paral-
lel applications require data inputs that must be trans-
fered with a high performance between data and com-
putational resources, transparently without any man-
ual intervention by the scientists.
Considering that e-Science infrastructures are ac-
cessible via Grid middleware and by scientists via
Grid clients, we identified two major extension re-
quirements for the support of database access for
biological applications in distributed environments.
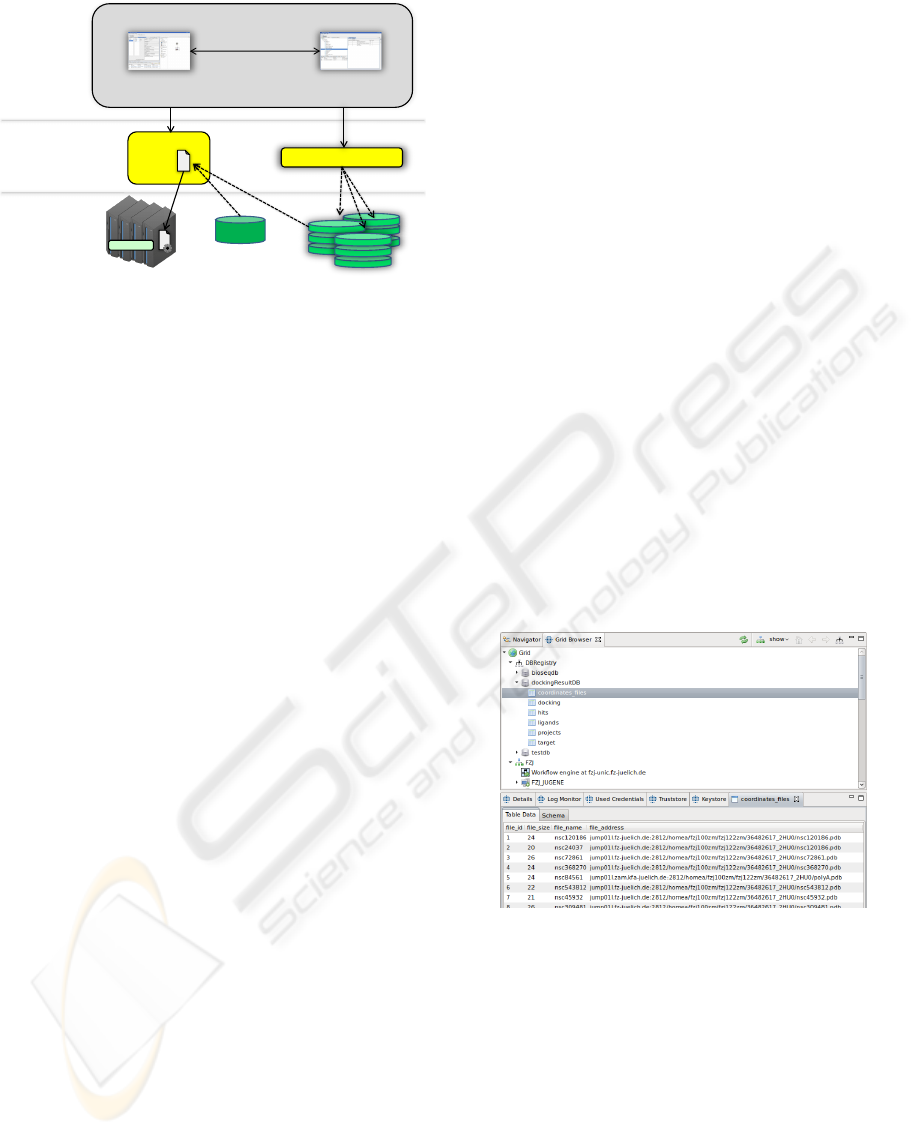
As shown in Figure 2, one demand is the access to
databases at the server tier. The other demand is the
support for graphical database access in our client ref-
erence implementation.
In this design, databases are integrated as Grid re-
sources on the target system tier. In fact, database
resources are mainly very heterogeneous on the phys-
ical and logical level, offering different data manage-
ment capabilities. Due to this, database access tools
typically provide standard-based interfaces such as
WS-DAIS OGF standard (Antonioletti et al., 2005)
to access database resources via common Grid pro-
tocols, e.g. Web services (Foster et al., 2004). As
the result of a review of available state-of-the-art
Grid data resource access tools, we integrated OGSA-
DAI (OGSA-DAI, 2008) in our UNICORE refer-
ence implementation for database access between the
server and target system tier. In our reference imple-
mentation we improved the client Java API of OGSA-
DAI, to integrate database access into the UNICORE
Rich Client (URC, 2009) (URC) with the objective
of realising the second demand of the database sup-
port. The URC was developed as a graphical client
for the UNICORE 6 middleware (Streit et al., 2009)
to support functionalities for accessing Grid resources
and services; many of them are related to the sub-
mission of jobs and workflows. The developed ex-
tension of the URC provides new basic Grid resource
types, which offer a comfortable new visual overview
BIOINFORMATICS 2010 - International Conference on Bioinformatics
204
MySQL
Application
WS-*
OGSA-DAI
Application Interface
Extension for browsing and
selecting data stored in
databases
Selected data
Supercomputer
Job
Description
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnfJlkfj
lkdsjfsndfldjvldjvndl
vdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnfJlkfj
lkdsjfsndfldjvldjvndl
vdlfnlkdvnf
Oracle
UNICORE
Client
Tier
Server
Tier
Targer System
Tier
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnfJlkfj
lkdsjfsndfldjvldjvndl
vdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnf
Jlkfjlkdsjfsndfldjvldj
vndlvdlfnlkdvnfJlkfj
lkdsjfsndfldjvldjvndl
vdlfnlkdvnf
GridFTP
Storage
UNICORE
Rich Client (URC)
Figure 2: The overview shows a typical three tier architec-
ture. Our development at the client tier enables users to
access databases and to chose data stored in databases as
application input within the URC.
of database resources. It also enables an intuitive view
to browse databases or view contents of tables or re-
sults of queries, as shown in Figure 3. Within this
scope, scientists can now filter the data for later reuse.
Another supporting service are SQL queries. Further-
more, selected data can automatically be delivered to
distributed computing resources for using it as data
input while processing jobs completely transparent to
the user.
While biological researchers mostly use the ap-
proach of storing metadata and a Grid file address in
the database, we additionally developed a wizard in-
cluding selection mechanism for database files with-
out the need of having any knowledge of SQL or the
like.
4 INTEROPERABILITY
APPLICATION
E-Science infrastructures, which mainly consist of
distributed resources and middleware that are well in-
terconnected, provide the computing power and data
resources required by complex biological problem
domains. The WISDOM [19] (World-wide In Sil-
ico Docking On Malaria) project effectively take ad-
vantage of such computational power by using a Grid
based two step bioinformatics virtual screening work-
flow in order to discover new drugs. The first part (A)
of the WISDOM workflow, as described in (Kasam
et al., 2009), is performed via molecular docking. In
WISDOM, molecular docking is performed on the
EGEE Grid infrastructure, which is the largest HTC-
driven Grid infrastructure in Europe.
The second part (B) of the workflow are molecu-
lar dynamic (MD) simulations. Therefore, the best
bindings of the molecular docking results of A are
refined by a complex MD simulation process. This
molecular dynamic simulation process (B) is realized
in WISDOM via the MD application package AM-
BER (AMBER, 2009). For the simulation of this sec-
ond part, researchers would like to use the URC AM-
BER support, as described in (Holl et al., 2009) and
the large-scale supercomputing facilities available on
DEISA, which is the European HPC-driven Grid in-
frastructure.
Typically the output of the molecular docking step
are stored in a MySQL database, which enables re-
searchers to conveniently use the developed database
access in the URC to filter these results by browsing
databases or creating specific queries. Additionally,
researchers can select the best promising outputs of
A for their usage as input for the next molecular dy-
namic refinement step (B). The submission and exe-
cution of the job as well as the data transfer is trans-
parent to the user, managed by the URC and the UNI-
CORE middleware. Hence, for the submission of
the WISDOM workflow to IIRM-based interoperable
infrastructures, scientists can access both, computa-
tional and data resources, within the URC, as in detail
described in (Holl, 2009). Another benefit is that long
running and manual data downloads and uploads are
avoided as well as the transfer of not necessary data.
Figure 3: The screenshot shows the Grid browser and the
table inside of a database.
The access to database resources as well as the in-
tegration of input data is standardized in the URC,
which causes an very identical functional usage for
various applications.
5 RELATED WORK
The execution of bioinformatics applications turned
out to be very effective in e-Science infrastructures.
Thus, the support of bioinformatics applications in
SUPPORTING SCIENTIFIC BIOLOGICAL APPLICATIONS WITH SEAMLESS DATABASE ACCESS IN
INTEROPERABLE E-SCIENCE INFRASTRUCTURES
205

e-Science infrastructures evolved in various scientific
projects, such as the Wildfire software (Tang et al.,
2005). It provides an integrated environment for con-
struction and execution of workflows over the com-
pute nodes of a cluster. But Wildfire only stores data
in a back-end database.
Another software tool is Pegasys (Shah et al.,
2004). It provides a GUI based workflow mecha-
nism with a unified data model to store results of pro-
grams. This uniform data model is a backend rela-
tional database management system, whereas our ap-
proach supports any database in Grid infrastructures.
To sum up, graphical tools in bioinformatics typi-
cally provide access to Grids or computational cluster
as execution environments. But foremost, they only
offer access to backend databases.
6 CONCLUSIONS
The support for biological applications via graphical
clients enables a centralized access to both, database
and computing resources in e-Science environments,
and is thus very important nowadays. The WIS-
DOM project takes advantage of an infrastructure
setup based on the IIRM and thus requires central-
ized access to both resources, to ease the evalua-
tion and selection process for input data that have to
be filtered for further refinement executions. There-
fore, the presented development enables researchers
to graphically browse the contents of database tables
and directly select inputs for the job execution as
well as submit the job via the UNICORE Rich Client.
Thereby, it avoids that researchers act on databases or
computational resources, which would instead require
low-level access and accounts on many different re-
sources. Furthermore, the manual download and up-
load steps for files are omitted, since input files are
automatically loaded into the job execution environ-
ment by the executing middleware system.
REFERENCES
AMBER (2009). Assisted Model Building with Energy Re-
finement. http://ambermd.org/.
Antonioletti, M. et al. (2005). Web services Data Ac-
cess and Integration - the Relational Realization (WS-
DAIR), Version 1.0. In Global Grid Forum (GGF).
Baldridge, K. and Bourne, P. E. (2003). The New Biology
and the Grid. John Wiley and Sons.
Berman, F., Fox, G. C., and Hey, A., editors (2003). Grid
Computing: Making The Global Infrastructure a Re-
ality. John Wiley and Sons.
D-Grid (2009). German National Grid. http://www.d-
grid.de.
DEISA (2009). Distributed European Infrastructure for Su-
percomputing Applications. http://www.deisa.org.
Ellert, M. et al. (2007). Advanced Resource Connector mid-
dleware for lightweight computational Grids. Future
Generation Computer Systems, (23):219–240.
Foster, I. et al. (2004). Modelling Stateful Resources with
Web Services Version1.1.
Foster, I. T. (2005). Globus Toolkit Version 4: Software
for Service-Oriented Systems. In NPC, volume 3779
of Lecture Notes in Computer Science, pages 2–13.
Springer.
gLite (2009). http://glite.web.cern.ch/glite/.
Holl, S. (2009). Eclipse-based client support for scien-
tific biological applications in e-science. Berichte des
Forschungszentrum Juelich. Juel-4303.
Holl, S. R. M., Demuth, B., Romberg, M., Streit, A., and
Kasam, V. (2009). Life science application support in
an interoperable e-science environment.
Kasam, V. et al. (2009). WISDOM-II: Screening against
multiple targets implicated in malaria using computa-
tional grid infrastructures. Malaria Journal, 8(1):88.
Laure, E. and Jones, B. (2008). Enabling Grids for e-
Science: The EGEE Project. CRC Press.
OGSA-DAI (2008). http://www.ogsadai.org.uk/.
Riedel, M. et al. (2008). Improving e-Science with Interop-
erability of the e-Infrastructures EGEE and DEISA.
In Proceedings of the 31st International Convention
MIPRO, Conference on Grid and Visualization Sys-
tems, pages 225–231.
Riedel, M. et al. (2009). Research Advances by using In-
teroperable e-Science Infrastructures - The Infrastruc-
ture Interoperability Reference Model applied in e-
Science. Cluster Computing, Special Issue Recent Ad-
vances in e-Science.
Shah, S. P. et al. (2004). Pegasys: software for execut-
ing and integrating analyses of biological sequences.
BMC Bioinformatics, 5.
Streit, A. et al. (2009). UNICORE 6 - A European Grid
Technology. IOS Press.
Tang, F. et al. (2005). Wildfire: distributed, Grid-enabled
workflow construction and execution. BMC Bioinfor-
matics, 6:69.
Taylor, I. J., Deelman, E., Gannon, D. B., and Shields, M.
(2006). Workflows for e-Science: Scientific Workflows
for Grids. Springer-Verlag New York, Inc.
URC (2009). UNICORE Rich Client.
http://www.unicore.eu/documentation/manuals/unico-
re6/files/RichClient.pdf.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
206
