
CLASSIFICATION OF MASS SPECTROMETRY DATA
Using Manifold and Supervised Distance Metric Learning
Qingzhong Liu
a,b
, Andrew H. Sung
a,b
, Bernardete M. Ribeiro
c,*
and Mengyu Qiao
a
a
Department of Computer Science,
b
Institute for Complex Additive Systems Analysis
New Mexico Tech, Socorro, NM 87801, U.S.A.
c
Department of Informatics Engineering, University of Coimbra, Portugal
Keywords: Proteomics, Mass spectrometry, Manifold, Distance metric learning, Classification, Support vector machine.
Abstract: Mass spectrometry becomes the most widely used measurement in proteomics research. The quality of the
feature set and applied learning classifier determine the reliability of the prediction of disease status. A well-
known approach is to combine peak detection and support vector machine recursive feature elimination
(SVMRFE). To compare the feature selection and to search for alternative learning classifier, in this paper,
we employ a distance metric learning to classification of proteomics mass spectrometry (MS) data.
Experimental results show that distance metric learning is promising for the classification of proteomics
data; the results are comparable to the best results by applying SVM to the SVMRFE feature sets. Results
also indicate that the good potential of manifold learning for feature reduction in MS data analysis.
1 INTRODUCTION
Mass spectrometry (MS), which includes a sequence
of the ratios of mass/charge (m/z), is currently used
for biomedical diagnosis and protein identification
(Petricoin and Liotta, 2003). The data mining
usually contains four steps, or pre-processing,
feature extraction, feature selection and
classification.
The objective of pre-processing is to purify the
data, and systematically represent the data for the
following steps. Generally speaking, MS data
contain two kinds of noise, electric noise and
chemical noise, which damage the classification
result. The chemical noise, usually shows as a
baseline along the spectrum, is generated by matrix
or ion overloading. Baseline correction computes the
local minimum value and draws a baseline
representing as the background noise and subtracts
the baseline from the spectrum. Williams et al
proposed a robust algorithm for computing the
baseline correction of MALDI-MS spectra
(Williams et al., 2005). The electronic noise, usually
randomly distributed in the spectra, is produced
from the electronic instrument. To remove the noise,
Chen et al designed a wavelet-based denoising
(Chen et al., 2007). The denoised data are
normalized to provide a systematic representation of
the spectra.
The next steps include extracting features from
the spectra, and forming the initial complete feature
set. The simplest way is to obtain every data point as
a discriminative feature, and finally have a huge
feature set including more than 15000 features (Li et
al., 2007), (Li et al., 2002), that is stupid for
classification. Recently, some researchers employed
an elaborated algorithm for peak detection and
performed a more aggressive feature extraction
(Coombes et al., 2007), (Hilario et al., 2006), (Shin
and Markey, 2006). A popular method to deal with
the feature selection in MS data classification is to
apply support vector machine recursive feature
elimination (SVMRFE) for getting a small subset of
peaks as input variables for the classification (Guyon
et al., 2002), (Vapnik, 1998), (Liu et al., 2008),
followed by SVM for the final testing.
To search for promising alternative classifiers
and to feature dimensionality reduction, in this
paper, we apply a distance metric learning, called
large margin nearest neighbor classifier (LMNN)
that is proposed by Weinberger (Weinberger et al.,
2006), to the classification of MS data. We also
compare the testing results on the reduced feature
sets with the use of a manifold learning.
Experimental results show that distance metric
learning is promising for the classification of
proteomics data; the results are comparable to the
best results by applying SVM to the SVMRFE
396
Liu Q., H. Sung A., M. Ribeiro B. and Qiao M. (2009).
CLASSIFICATION OF MASS SPECTROMETRY DATA - Using Manifold and Supervised Distance Metric Learning .
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing, pages 396-401
DOI: 10.5220/0001556403960401
Copyright
c
SciTePress

feature sets. Results also indicate that the good
potential of manifold learning for feature reduction
in MS data analysis.
The remainder is organized as follows. Pre-
processing algorithms are briefly discussed in
section two, LMNN is introduced in section three
and experimental results are given in section four,
followed by our conclusions in section five.
2 PREPROCESSING DATA
MS data have high dimensionality and small number
of samples. Both chemical and electrical noises are
involved in the signal. The redundancy of the
spectra, different reference points, and unaligned
feature points increase the computational intensity
and decrease the classification accuracy. To deal
with these issues, the proposed processing
procedures include spectra re-sampling, wavelet de-
noising, baseline correction, normalization, peak
detection and alignment.
2.1 Spectra Re-sampling and Wavelet
De-noising
The mass spectrum data is in a discrete format and
the intervals are not equal in the whole spectrum.
For high-resolution data, the high-frequent noise and
redundant data points harm the quality of the dataset.
So, we have to set the common low-frequent mass
value to every sample spectrum in order to give a
unified representation. By using spline interpolation,
we resample the data and confine the interval to a
unified size. Before re-sampling, the sample
spectrum has a little variation from the true
spectrum. The data is re-sampled to a standard
discrete data which could be analyzed in frequency
domain. The electrical noise is generated during the
mass spectrum acquisition by the instrument and it is
almost random distributed noise. The next step is to
employ discrete wavelet transform for eliminating
the electrical noise. By applying a wavelet
transform, the original signal is decomposed into
multi-level wavelet coefficients. By setting up a
threshold value, given percentiles of lower value
coefficients are removed. Then, we apply
polynomial filter of second order to smooth the
signal and get better data quality.
2.2 Baseline Correction and
Normalization
To minimize the chemical noise, the baseline should
be subtracted from the spectrum. In order to obtain
the baseline, the local minima should be computed
by assigning an appropriate window size. Then, we
use spline interpolation to fit the baseline. In order to
compare sample spectra, we need to normalize the
data to represent the data in a systematic scale.
2.3 Peak Detection and Qualification
The final feature acquisition of MS data is to obtain
the peak position and its magnitude. In our mass
spectrum experiment, the peak detection method
proposed by Coombes et al (Coombes et al, 2005) is
performed on mean spectrum rather than individual
spectra, and we used the ad hoc method based on
signal to noise ratio to select the large peaks
(Coombes et al., 2007).
3 DISTANCE METRIC
LEARNING
Distance metric learning includes supervised
learning and unsupervised learning.
For unsupervised distance metric learning or
called manifold learning, the main idea is to learn an
underlying low-dimensional manifold where
geometric relationship between most of the observed
data are preserved. Every dimension reduction
approach is essentially to learn a distance metric
without label information. Manifold learning
algorithms can be divided into global linear
dimension reduction approaches, including Principle
Component Analysis (PCA) and Multiple
Dimension Scaling (MDS), global nonlinear
approaches, for instance, ISOMAP (Tenenbaum et
al., 2000), local linear approaches, including Locally
Linear Embedding (LLE) (Saul and Roweis, 2003)
and the Laplacian Eigenmap (Belkin and Niyogi,
2003). Here we introduce a locally linear embedding
proposed in Roweis and Lawrance, 2000) that is a
local method to establish the mapping relationship
between the observed data and the corresponding
low-dimensional data and to preserve local order
relation of data in both the embedding space and the
intrinsic space, described as follows:
1. Find the n nearest neighbor for each x
i
in
the dataset. By assuming that each data
point and its neighbors lie on a locally
linear patch of the manifold, the local
geometry of these patches can be
characterized by linear coefficients that
reconstruct each data point from its
neighbors. The reconstruction error can be
measured by
CLASSIFICATION OF MASS SPECTROMETRY DATA - Using Manifold and Supervised Distance Metric Learning
397

2
(W) || W ||
iijij
j
x
x
ϕ
=−
∑
(1)
2. Construct the approximation matrix by
minimizing
1
|| W ||
nn
iijij
ij
x
x
=
−
∑∑
(2)
s.b.
W1
ij
j
=
∑
.
3. Construct a neighbor-preserving mapping.
The idea is that the reconstruction weights
reflect intrinsic geometric properties of the
data and are invariant to the linear
transform from a high dimensional
coordinates of each neighborhood to global
internal coordinates on a low dimensional
manifold. The details are given in Roweis
and Lawrance, 2000).
Supervised distance metric learning can be
divided into global distance metric learning and
local distance metric learning. The global learns the
distance metric in a global sense, i.e., to satisfy all
the pairwise constraints. The local approach seeks
only to meet local pairwise constraints.
In supervised global distance metric learning, the
representative work is to formulate distance metric
learning as a constrained convex programming
problem (Xing et al., 2003). It learns a global metric
distance that minimizes the distance between the
data pairs in the equivalence constraints subject to
the constraint that the data pairs in the inequivalence
constraints are well separated. In local adaptive
distance metric learning, many researchers presented
approaches to learn appropriate distance metric to
improve KNN classifier (Domeniconi and
Gunopulos, 2002), (Peng et al., 2002), (Goldberger
et al., 2005), (Zhang et al., 2003), (Zhang et al.,
2005).
Inspired by the work on neighborhood
component analysis (Goldberger et al., 2005) and
metric learning by energy-based models (Chopra et
al., 2005), Weinberger et al. proposed a distance
metric learning for Large Margin Nearest Neighbor
classification (LMNN). Specifically, the
Mahanalobis distance is optimized with the goal that
k-nearest neighbors always belong to the same class
while examples from different classes are separated
by a large margin (Weinberger et al., 2006). LMNN
has several parallels to learning in support vector
machines (SVMs), for example, the goal of margin
maximization and a convex objective function based
on the hinge loss. In multi-classification, the training
time of SVMs scales at least linearly in the number
of classes, by contrast, LMNN has no explicit
dependence on the number of classes (Weinberger et
al., 2006). We introduce the idea of LMNN as
follows:
Given a training set of n labeled samples and the
corresponding class labels
{}
1
,
n
ii
i
xy
=
, the binary
matrix
{0,1}
ij
y ∈
indicates whether or not the
labels
i
y
and
j
y
match. And
{0,1}
ij
η
∈
indicates
whether
j
x
is a target neighbor of
i
x
. Both matrices
ij
y
and
ij
η
are fixed during training. The goal is to
learn a linear transformation
L: R R
d
→
that
optimizes KNN classification. The transform is used
to compute squared distance as
2
D(,)||L(,)||
ij ij
xx xx=
(3)
The cost function is given as follows:
2
2
2
(L) L( )
(1 ) 1 L ( ) L( )
ij i j
ij
ij jl i j i l
ijl
xx
Cy xxxx
εη
η
+
=−
⎡
⎤
+−+−−−
⎢
⎥
⎣
⎦
∑
∑
(4)
Where [z]
+
= max(z,0) denotes the standard
hinge loss and the constant C > 0. The first term
penalizes large distances between each input and its
target neighbors and the second term penalizes small
distances between each input and all other inputs
that do not share the same label.
The optimization of eq. (2) can be reformulated
as an instance of semidefinite programming (SDP)
Vandenberghe and Boyd, 1996) and the global
minimum of eq. (2) can be efficiently computed.
Mahalanobis distance metric
MLL
T
= , then eq. (1)
is
D( , ) ( ) M( )
T
ij i j i j
x
xxx xx=− −
(5)
Slack variables
ij
ξ
for all pairs of differently
labeled inputs are introduced so that the hinge loss
can be mimicked. The resulting SDP is given by:
Minimize
(L) ( ) ( ) (1 )
T
ij i j i j ij jl ijl
ij ijl
xxMxx C y
ε
ηηξ
=− −+ −
∑
∑
Subject to
(1)
()M()( )M( )1
il il i j i j ijl
xx xx xx xx
ξ
−
−−− − ≥−
(2)
0
ijl
ξ
≥
(3)
M0≥
BIOSIGNALS 2009 - International Conference on Bio-inspired Systems and Signal Processing
398
4 MATERIALS AND
EXPERIMENTS
4.1 Dataset
The following two mass spectrometry datasets have
been tested in our experiment.
1. High resolution time-of-flight (TOF) mass
spectrometry (MS) proteomics data set from surface-
enhanced laser/desorption ionization (SELDI)
ProteinChip arrays on 121 ovarian cancer cases and
95 controls. The data sources can be accessed by
FDA-NCI Clinical Proteomics at
http://home.ccr.cancer.gov/ncifdaproteomics/ppatt
erns.asp
2. The breast cancer QC SELDI spectra data set
was studied by Pusztai et al. (Pusztai et al., 2004).
Here we utilized data of 57 controls and 51 cases.
The data set may be downloaded at:
http://bioinformatics.mdanderson.org/Supplemen
ts/Datasets
We process the dataset according to the methods
described in section 2 for peak detection. And then
apply LMNN to the detected peak spectra data.
Since SVMRFE is a classical feature selection based
on the weights of the support vectors (Guyon et al.,
2002) and is widely used in the classification of
proteomics data with the use of SVM, we also
compare the results by using LMNN with Euclidean
distance, Mahalanobis distance, and energy-based
classification (Weinberger et al., 2006) and SVM
combining with SVMRFE on the detected peak data.
In each experiment, 80% samples are randomly
chosen for training and the remaining 20% samples
are tested. We repeated the experiments 10 - 100
times and compared the average testing results.
Besides applying SVMRFE to the features
chosen by peak-detection algorithm, we also apply
the manifold learning to the features chosen by
peak-detection and the features filtered by rank-sum
test without peak-detection, and obtain the reduced
features that are mapped from high-dimension to
low-dimension, then we apply a support vector
machine and KNN to the reduced feature sets and
compare the testing results.
4.2 Experiments on Peak Detection
with SVMRFE
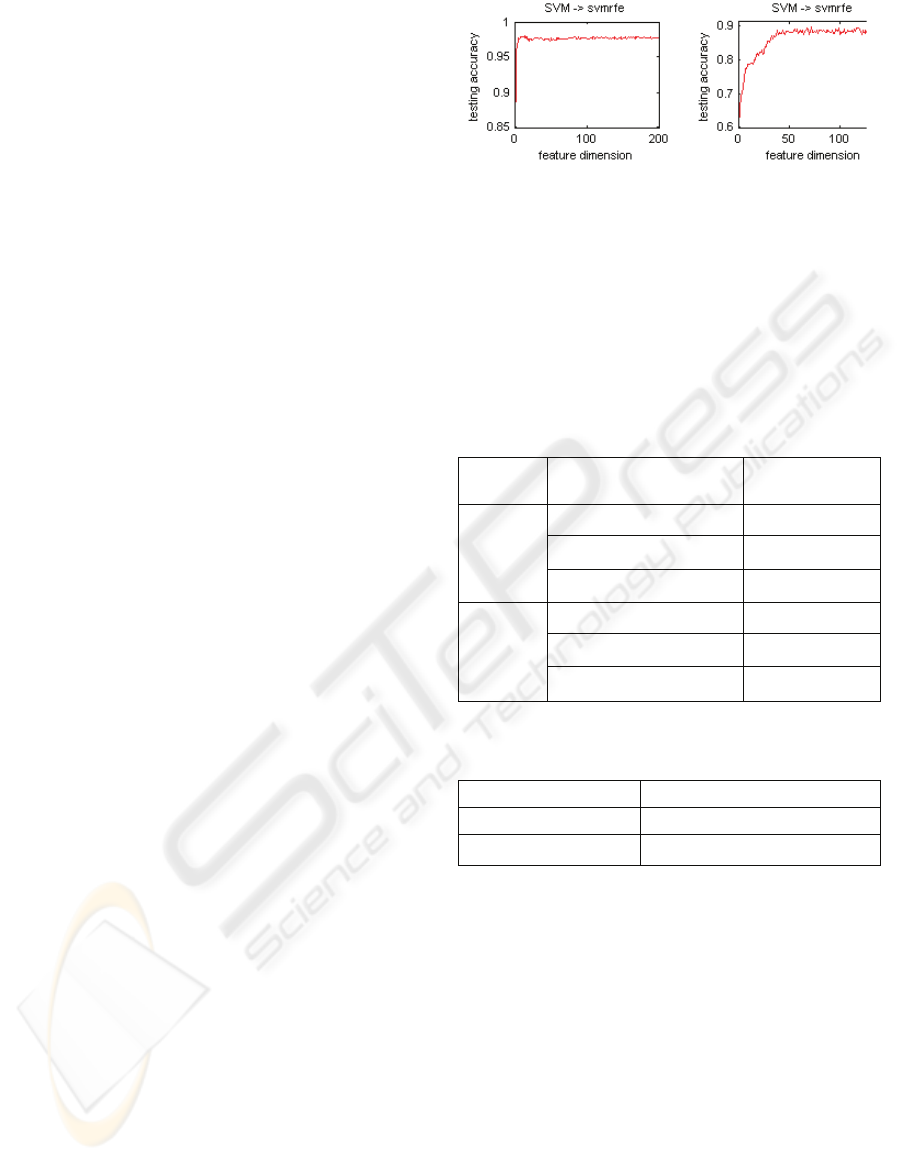
Figure 1 list the average testing accuracy values by
applying SVM to SVMRFE on ovarian cancer data
set and breast cancer data set, respectively. These
data sets were preprocessed with the use of peak-
detection algorithms before the use of SVMRFE.
(a) Ovarian cancer (b) Breast cancer
Figure 1: The testing accuracy by applying SVM to the
feature sets with the use of SVMRFE on ovarian cancer
data set (a) and breast cancer data set (b).
Table 1 lists the average testing accuracy by
applying LMNN classifiers to the peak data sets.
Table 2 lists the best averaging testing accuracy by
applying SVM to the feature sets chosen by
SVMRFE from the peak data sets.
Table 1: Average testing accuracy with LMNN classifiers.
Data set Classifier
Testing
accuracy
Ovarian
cancer
LMNN_energy
99.3%
LMNN_Euclidean 84.6%
LMNN_Mahalanobis 99.0%
Breast
cancer
LMNN_energy 81.8%
LMNN_Euclidean 84.6%
LMNN_Mahalanobis 81.7%
Table 2: The best average testing accuracy by applying
SVM to feature sets ranked by SVMRFE.
Data set Testing accuracy
Ovarian cancer 98.0%
Breast cancer
89.5%
Comparing the results listed in tables 1 and 2, we
can see that, although the testing results in table 1
are not as good as the best result by applying SVM
to SVMRFE feature sets for breast cancer data set,
the results by applying LMNN classifiers based on
energy classification and Mahalanobis distance are
both better than the best result obtained by applying
SVM to SVMRFE feature sets. It indicates that
LMNN classifiers with energy classification and
Mahalanobis metric are competitive for the
classification of proteomics data, especially
considering that the best testing accuracy by
applying SVM to SVMRFE feature sets after we
tested each dimensionality from 1 to 200 for ovarian
cancer data set and tested each dimensionality from
1 to 150 for breast cancer data set. Furthermore,
normally we cannot accurately predict the best
CLASSIFICATION OF MASS SPECTROMETRY DATA - Using Manifold and Supervised Distance Metric Learning
399
testing results that correspond to which
dimensionality.
We also test the classification results by applying
LMNN to the feature sets chosen by SVMRFE, and
compare the best testing result and the least number
of the features corresponding to the best result in
each experiment against the results with the use of
SVM, shown in table 3. Experimental results
indicate comparable results by applying LMNN and
support vector machines.
Table 3: The highest testing accuracy and the least number
of the features corresponding to the highest testing results.
Data
set
Classifier
Testing
accuracy
Feature
number
Ovaria
n
cancer
LMNN_energy
99.5%
14
LMNN_Euclidean 98.4% 10
LMNN_Mahalanobis
99.5%
15
SVM 99.3% 11
Breast
cancer
LMNN_energy 92.6% 27
LMNN_Euclidean 90% 28
LMNN_Mahalanobis 92.5% 25
SVM
94%
68
4.3 Manifold for Feature Reduction
We also preprocess the data sets by using peak-
detection algorithms (method one) and filtered the
data sets by using rank sum test without peak-
detection (method two), then we apply the manifold
learning Roweis and Lawrance, 2000) to reduce the
features, finally we employ a SVM and a KNN to
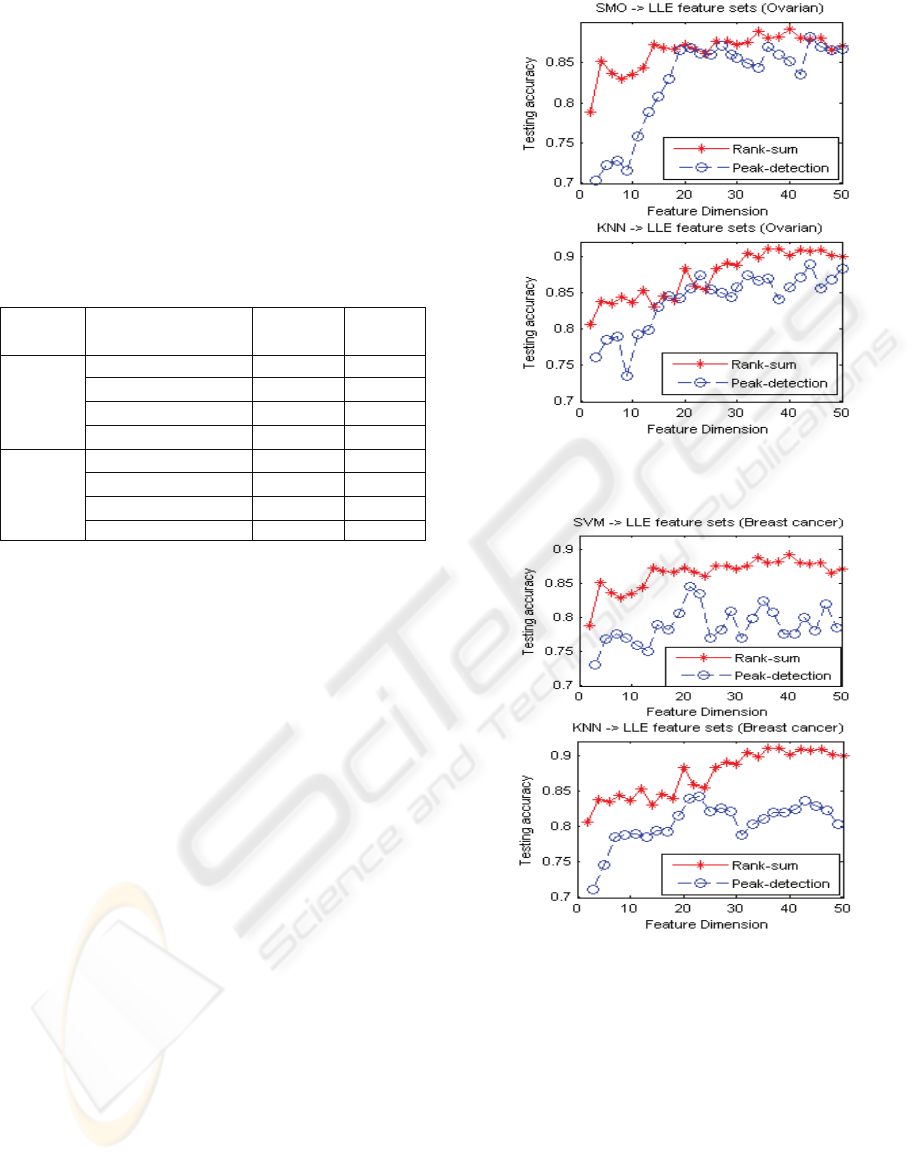
classify the data sets. Figure 2 gives the comparison
of the testing accuracy of these two methods, with
and without peak-detection on the ovarian cancer
data. Figure 3 plots the testing results on breast
cancer data.
Figures 2 and 3 indicate that the testing results
on the reduced feature sets preprocessed from rank
sum test are better than the results on the reduced
feature sets preprocessed by peak detection. It seems
to imply that peak detection cannot include all useful
features. This is worthy of further study.
We also note that the testing results on ovarian
data set are not better than the results obtained by
using SVMRFE, shown in 4.2. However, the results
on breast cancer are comparable to the results with
the use of SVMRFE, given in section 4.2. It
indicates that unsupervised metric learning or
manifold learning holds good promise in dealing
with high-feature dimension data for dimension
reduction and this makes interesting future study.
Figure 2: The testing results of the LLE reduce feature sets
on peak-detection and rank sum test, ovarian cancer data.
Figure 3: The testing results of the LLE reduce feature sets
on peak-detection and rank sum test, breast cancer data.
5 CONCLUSIONS
In this paper, we compared a supervised distance
metric learning, large margin nearest neighbor
classifier and SVM for classification of mass
spectrometry proteomics data. Experiments
produced good results of applying distance metric
learning to proteomics data, comparable to the
results by applying SVM. Our results also indicate
the potential of manifold learning in feature
BIOSIGNALS 2009 - International Conference on Bio-inspired Systems and Signal Processing
400

reduction. Further, our results also indicate that,
peak detection may not be the optimal choice for
pre-processing proteomics data.
ACKNOWLEDGEMENTS
Support for this research received from the Institute
of Complex Additive Systems Analysis, a unit of
New Mexico Tech, is gratefully acknowledged.
REFERENCES
Petricoin, E. and Liotta, L. (2003), Mass spectrometry-
based diagnostic: the upcoming revolution in disease
detection. Clin. Chem., 49, pp.533-534.
Williams, B., Cornett, S., Dawant, B., Crecelius, A.,
Bodenheimer, B. and Caprioli, R. (2005), An
algorithm for baseline correction of MALDI mass
spectra, Proceedings of the 43rd annual Southeast
regional conference, March 18-20, 2005, Kennesaw,
Georgia.
Chen, S., Hong, D. and Shyr, Y. (2007), Wavelet-based
procedures for proteomic mass spectrometry data
processing, Computational Statistics & Data Analysis,
2007, Vol. 52, issue 1, pp.211-220.
Li, L. et al. (2004), Applications of the GA/KNN method
to SELDI proteomics data. Bioinformatics, 20,
pp.1638-1640.
Petricoin, E. et al. (2002), Use of proteomics patterns in
serum to identify ovarian cancer. The Lancet, 359,
pp.572-577.
Coombes, K. et al. (2007), Pre-processing mass
spectrometry data. In Dubitzky, M., et al. (eds.),
Fundamentals of Data Mining in Genomics and
Proteomics. Kluwer, Boston, pp.79-99.
Hilario, M. et al. (2006), Processing and classification of
protein mass spectra. Mass Spectrom. Rev., 25:409-
449.
Shin, H. and Markey, M. (2006), A machine learning
perspective on the development of clinical decision
support systems utilizing mass spectra of blood
samples. J. Biomed. Inform. 39, pp.227-248.
Furey, T. et al. (2000), Support vector machine
classification and validation of cancer tissue samples
using microarray expression data. Bioinformatics, 16:
906-914.
Coombes, K. et al. (2005), Improved peak detection and
quantification of mass spectrometry data acquired
from surface-enhanced laser desorption and ionization
by denoising spectra with the undecimated discrete
wavelet transform, Proteomics, Volume 5, Issue 16.
Duan, K. and Rajapakse, J.C. (2004), SVM-RFE peak
selection for cancer classification with mass
spectrometry data. APBC 2005: pp.191-200.
Guyon, I., Weston, J., Barnhill, S. and Vapnik, V.N.
(2002), Gene Selection for Cancer Classification using
Support Vector Machines. Machine Learning. 2002
46(1-3): pp.389-422.
Vapnik,V.N. (1998), Statistical Learning Theory. John
Wiley and Sons, New York.
Brown, M.P.S. et al. (2000), Knowledge-based analysis of
microarray gene expression data by using support
vector machines. Pro. Nat Acad. Sci., 97, pp.262-267.
Liu, Q., Sung, A.H., Chen, Z. and Xu, J. (2008), Feature
Mining and Pattern Classification for Steganalysis of
LSB Matching Steganography in Grayscale Images,
Pattern Recognition, 41(1): pp.56-66.
Tenenbaum, J., Silva, V. de and Langford, J. C. (2000), A
global geometric framework for nonlinear
dimensionality reduction, Science, vol. 290, pp.2319-
2323.
Saul, L. K. and Roweis, S. T. (2003), Think globally, fit
locally: Unsupervised learning of low dimensional
manifolds, Journal of Machine Learning Research,
vol. 4, pp.119-155.
Belkin, M. and Niyogi, P. (2003), Laplacian eigenmaps
for dimensionality reduction and data representation,
Neural Computation, 15( 6):1373-1396.
Xing, E., Ng, A., Jordan, M., and Russell, S. (2003),
Distance metric learning with application to clustering
with side-information, in Proc. NIPS, 2003.
Domeniconi, C. and Gunopulos, D. (2002), Adaptive
nearest neighbor classification using support vector
machines, Proc. NIPS, 2002.
Peng, J., Heisterkamp, D. and Dai, H. (2002), Adaptive
kernel metric nearest neighbor classification, Proc.
International Conference on Pattern Recognition,
2002.
Goldberger, J., Roweis, S., Hinton, G. and Salakhutdinov,
R. (2005), Neighbourhood components analysis, in
Proc. NIPS, 2005.
Zhang, Z., Kwok, J. and Yeung, D. (2003), Parametric
distance metric learning with label information, in
Proc. International Joint Conference on Artificial
Intelligence, 2003.
Zhang, K., Tang, M. and Kwok, J. T. (2005), Applying
neighborhood consistency for fast clustering and
kernel density estimation. in Proc. Computer Vision
and Pattern Recognition, 2005, pp. 1001-1007
Chopra, S., Hadsell, R. and. LeCun Y. (2005), Learning a
Similarity Metric Discriminatively, with Application
to Face Verification, Proc. Computer Vision and
Pattern Recognition, 2005, Vol. 1, pp.539-546.
Weinberger, K., Blitzer, J. and Saul, L. (2006), Distance
metric learning for large margin nearest neighbor
classification, in Proc. NIPS, 2006, pp.1475-1482.
Pusztai et al. (2004), Pharmacoproteomic Analysis of
Prechemotherapy and Postchemotherapy Plasma
Samples from Patients Receiving Neoadjuvant or
Adjuvant Chemotherapy for Breast Carcinoma,
Cancer 100: pp.1814-1822.
Vandenberghe, L. and Boyd, S.P. (1996), Semidefinite
programming, SIAM Review, 38(1): 49-95.
Roweis, S. T. and Lawrance, K. S. (2000), Nonlinear
dimensionality reduction by locally linear embedding,
in Science, vol. 290, 2000, pp.2323-2326.
CLASSIFICATION OF MASS SPECTROMETRY DATA - Using Manifold and Supervised Distance Metric Learning
401
