
A NOVEL EVOLUTIONARY FRAMEWORK FOR FEATURE
MATCHING
∗
Biao Wang and Chaoying Tang
College of Automation Engineering, Nanjing University of Aeronautics and Astronautics, Yudao Street, Nanjing, China
Keywords:
Feature Matching, Queen-bee Evolution, Genetic Operators.
Abstract:
The paper presents a new feature matching scheme based on the Queen-bee Evolution for two uncalibrated
images. Matching features needs an exhaustive search in a vast space, for which evolutionary algorithms
are recommended recently. This paper propose a simple and effective algorithm. We intuitively encode a
string of integer numbers assigned to the features as chromosomes and develop a novel crossover operator
respectively which can preserve the position information without any disruption. We also tailor swap mutation
operator to prevent from premature convergence and invalid solutions. As a result, the proposed algorithm can
quickly achieve the global or near global optimal solution cooperating with the linear ranking selection and
the elitist replacement. Meanwhile, it is a more general framework for matching various types of features. The
experimental results illustrate the performance of the proposed approach.
1 INTRODUCTION
Matching features between two uncalibrated images
constitutes a fundamental step in a variety of com-
puter vision applications, including automatic robot
navigation, target recognition, motion estimation, etc.
Although solutions to the problem have been explored
for many years, it remains of central interest because
no general method can be proposed and the focus on
the matching process has to vary with the require-
ments of different applications. We intent to esti-
mate the pose of an aircraft in air by vision techniques
and some classical matching algorithms, such as re-
laxation, cross-correlation, Least-Median-of-Squares
were tested. Unfortunately all of them are not robust
enough in our case. We make our first effort here al-
though the result is not perfect now.
Feature matching is usually cast as a combinato-
rial optimization problem and the search strategy in-
volved is one of the important parts to achieve optimal
solutions. As a search strategy, Genetic Algorithms
(GAs) take more attention recently due to its advan-
tages: the global search ability, intrinsically parallel
computing, the insensitivity to initial values, effec-
tive search on vast solution spaces, etc. (Chai and
Ma, 1998) match corner points extracted from two
∗
Supported by National Natural Science Foundation of
China (No. 60674100)
uncalibrated images using an evolutionary framework
and propose a 2D chromosome structure with binary
entries and an adaptation operator. (Ruichek et al.,
2000) use the same chromosome structure and give
the feature matching scheme for the images taken by
linear stereo cameras. (Beveridge et al., 2000) com-
pare three line matching techniques and recommend
the method based on a Messy GA. Also, other evolu-
tionary frameworks are used to search for correspond-
ing features (Yuan et al., 2004).
This paper proposes a novel GA-based matching
scheme based on the Queen-bee Evolution (QE) to
establish correspondences between two uncalibrated
images. Although in our specific implementation cor-
ners are used as features, it is indeed a relatively
more general framework for matching various feature
types, such as lines, edges, and so on. Benefiting
from a novel chromosomal encoding with its proper
crossover and mutation operators we developed, the
proposed algorithm is simple and effective. Section
2 introduces the QE briefly. Section 3 describes our
QE-based matching scheme in detail. Section 4 gives
the experimental results to illustrate the performance
of our approach. Section 5 concludes our work.
641
Wang B. and Tang C. (2008).
A NOVEL EVOLUTIONARY FRAMEWORK FOR FEATURE MATCHING.
In Proceedings of the Third International Conference on Computer Vision Theory and Applications, pages 641-644
DOI: 10.5220/0001075606410644
Copyright
c
SciTePress

2 QUEEN-BEE EVOLUTION
Queen-bee evolution is really a variation of classic
GAs. It mimics the nature that the queen bee cross-
breeds with drone bees and plays a major role in the
reproduction process. Here, the queen is the fittest
individual in a generation, and the drones are the in-
dividuals selected as parents to crossbreed with the
queen. It is noticeable that all drones crossbreed only
with the queen-bee but not with each other. This en-
hances the exploitation of genetic information in the
queen, meanwhile leading to an increased probabil-
ity of premature convergence. To solve the problem,
some individuals in a population are strongly mu-
tated so that the exploration of GAs is reinforced. In-
terested readers are referred to the reference (Jung,
2003) for detailed information.
3 MATCHING WITH QUEEN-BEE
EVOLUTION
The purpose of feature matching is to establish corre-
spondences between the features of two images taken
by an uncalibrated camera system. We use corner
points as features because they are easily found in
man-made scenes. Assume that they have been ex-
tracted with an improved Harris corner detector (Har-
ris and Stephens, 1988; Schmid et al., 2000) from the
two images independently. The following will per-
form the matching task benefiting from QE.
3.1 Chromosomal Encoding of
Solutions
Feature matching can be cast as such a combinato-
rial optimization problem that the M features in the
first image match to the N features in the second im-
age. That is, a candidate solution of the problem is
a specific mapping between the two feature sets. Ac-
cording to the uniqueness assumption that any feature
in one image can be assigned to at most one feature
in the other, permutation is preferable to encode the
feasible solutions since it has been widely used to
solve similar problems, such as TSP, JSP, and QAP
(Bierwirh et al., 1996). However, two main differ-
ences need be considered: feature matching focuses
on the mapping between the elements of two feature
sets but not on the order of the elements in one set; and
usually some features in one image may not match to
any feature in the other, hence a correct solution often
contains only a portion of all extracted features.
We define the chromosome as an integer string
with length M, each position corresponding to a fea-
ture in the first image (Fig. 1). The integer values
ranging from 1 to N on those positions are the la-
bel numbers uniquely assigned to the features in the
second image. They are named talking genes against
the value 0’s, so called dummy genes, which means
no match in the second image. In a specific chromo-
some, each talking gene is unique whereas a dummy
gene often duplicates several times because multiple
features in the first image usually don’t match to any
feature in the second. Inversely, multiple features in
the second image usually don’t match to any feature
in the first, so the genes in a chromosome constitute
only a k-subset of all available talking genes {1, 2, . . .,
N}. That is, the encoding is not a true permutation, so
called partial permutation.
Figure 1: A chromosome with the partial permutation en-
coding.
3.2 Matching Constraints and Fitness
Function
On the view of optimization, our solution scheme is to
find the permutation of the feature labels in the second
image that maximizes a given fitness function. We
define the fitness of chromosomes as follows.
First, the normalized cross-correlation is com-
puted for the neighborhood of every point in the first
image to the ones in the second. Combining with spa-
cial similarity of features, we threshold the correlation
score to reduce the search space, which is also em-
ployed in the initialization and the mutation phases of
our evolution process.
Second, the neighbor features are found for ev-
ery candidate match containing in a chromosome. If
its neighbors are also candidate matches in the chro-
mosome, they give support for each other and have
higher matching strength. Detailed computing is re-
ferred to (Zhang et al., 1994). Differently, the rotation
limitation is not used here. Furthermore, the unique-
ness assumption has been embedded in our encod-
ing process so that the computing symmetry problem
does not exist here.
Third, the fitness function summarizes the
strengths of all candidate matches in the chromosome.
3.3 Selection and Replacement
Selection is applied to population of each genera-
tion so that fitter chromosomes, i.e. those satisfy the
matching constraints better, will have more breeding
VISAPP 2008 - International Conference on Computer Vision Theory and Applications
642
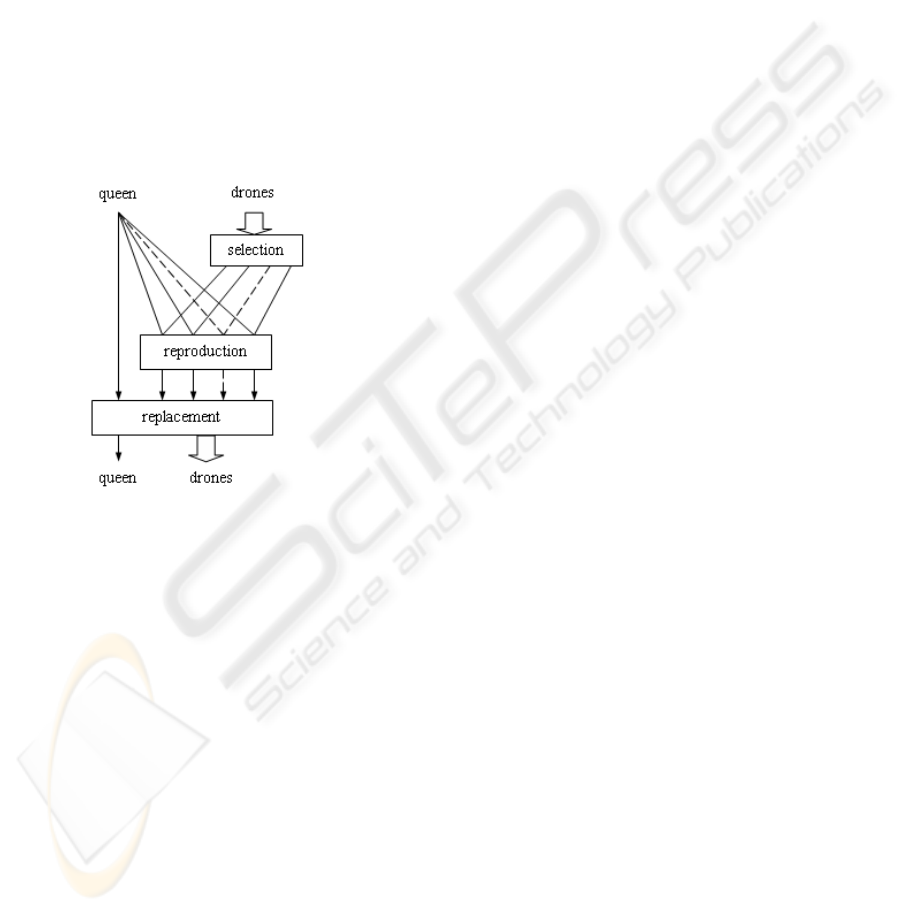
chances to have offspring by crossover. Linear rank-
ing selection (Zhang and Kim, 2000) is proved to
be efficient in our proceeding experiments. Differ-
ent from simple GAs, it only operates on the drone
bees and the queen-bee in QE must be one of the two
parents to crossover (Fig. 2).
Replacement is a process that chooses survivors
from offspring to form the next generation. A widely
used scheme replaces the current population with its
offspring no matter whether they are fitter than their
parents. Slightly different, we mix the old queen with
the offspring first. Then, a new queen is picked out
and other offspring is preserved as drones for the next
generation. Hence, the old queen may survive with-
out crossoverand the fittest chromosomewill neverbe
kicked out of population during evolution, so called
Elitist Replacement.
Figure 2: Selection and replacement.
3.4 Crossover Operator
The basic idea of the crossover is that several char-
acteristics of some genotypes in parents are recom-
bined into their offspring. With permutation encod-
ing, a gene in a chromosome contains the informa-
tion of position, order, and/or adjacency(Starkweather
et al., 1991). Various crossover operators have been
developed intending to preserve respective favorable
information depending on problems. In our problem,
we expect the position to be of particular interest be-
cause it directly expresses corresponding relations be-
tween two feature sets. Cycle Crossover is such an
operator that always preserves the absolute position
of genes from one parent or the other without any dis-
ruption. Unfortunately, the partial permutation may
present multiple dummy genes in one chromosome,
and all genes in the chromosome usually constitute
only a subset of all available genes so that the cycle
crossover doesn’t respect the semantic properties of
the representation.
We define a new crossover for the partial permu-
tation encoding. Firstly, some positions are randomly
selected in the parents to crossover, and the genes on
those positions transfer to their two offspring at the
same positions. The genes on the other positions in
the offspring are inherited from the other parent at
the same positions. Secondly, duplicate talking genes
are detected. If two genes present identically in the
same offspring, one of the genes cannot be inherited
from its original parent but from the other. The sec-
ond operation is repeated until no duplicate talking
genes present in the offspring. Obviously, two differ-
ent offspring can be obtained from the process above
at the same time and they are of genetic complement,
so called complementary crossover. Thus, position
information is preserved without any disruption.
3.5 Mutation Operator
Mutation injects new genes into population to pre-
vent GAs from premature convergence. (Brizuela
and Aceves, 2003) compared three types of mutation
operators for multi-objective flowshop optimization
problems using permutation codes. They are all less
effective to the partial permutation, even produce in-
valid solutions. We prefer to tailor the swap operator
to our use.
Before mutation, a probability check for mutation
is performed on every position in a chromosome. If
the probability check is passed on a certain position,
the gene on that position will be mutated to an al-
ternative one randomly selected in the subset of po-
tential genes for the position. Here, a potential gene
means satisfied with intensity and spacial similarities
between the two corresponding features. If the alter-
native gene isn’t present in the same chromosome, a
new gene is injected into the current chromosome. If
the alternative gene is found on another position in
the chromosome except for dummy genes and they
are swappable, the two genes are swapped one time
between the two positions; otherwise a dummy gene
would be injected into the chromosome. As a result,
the tailored swap mutation can add a new gene into
a chromosome, swap two genes, or delete an existent
talking gene from the chromosome.
It is worth noting that QE with normal mutation
probability is prone to premature convergence be-
cause it exploits the genetic information of the queen
too intensively during reproduction processes. To
solve the problem, two mutation probabilities are em-
ployed in QE. Some chromosomes are normally mu-
tated as in the simple GAs and the others are strongly
mutated. The better values set for the normal mu-
A NOVEL EVOLUTIONARY FRAMEWORK FOR FEATURE MATCHING
643

tation rate, normal and strong mutation probabilities
have to be determined in experiments.
4 EXPERIMENTAL RESULTS
As shown in Fig. 3, corners extracted independently
from the two uncalibrated images are denoted by sym-
bol ”+” and ”x”, respectively. There are 34 points in
the first image and 37 in the second. In either image,
there are some corners without matches in the other.
The proposed GA is set as follows: population
size 20, crossover probability 1.0, normal mutation
probability 0.01, strong mutation probability 0.3, and
normal mutation rate 0.5. Among them, the later two
are the QE-specific parameters.
The generations needed for convergence are 50
averagely that means the speed of convergence is fast.
Certainly, it varies with the different number of fea-
tures, parameter settings, and so on. The resultant cor-
respondences are shown in the same figure denoted
by symbol ”*” assigned with a number. Observably,
the percentage of matched points achieves nearly the
maximum.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
Figure 3: The resultant correspondences.
5 CONCLUSIONS
The paper has presented a novel feature-based match-
ing scheme using queen-bee evolution. Intuitively, the
candidate solutions to correspondences of two uncal-
ibrated images are encoded with the label numbers of
features. Respectively, a new crossover is developed
to preserve the position information without any dis-
ruption, and the swap mutation is improved to respect
the semantic properties of the genetic representation.
The matching scheme uses the measure very similar
in the form to that used in (Zhang et al., 1994), but
a modified version. Comparing with the relaxation
technique, our approach can obtain more correct cor-
respondences and achieve the global or near global
optimal solution more easily. The experiment shows
that it gets convergence quickly and isn’t sensitive to
the initial values with proper selection and replace-
ment techniques.
REFERENCES
Beveridge, J. R., Balasubramaniam, K., and Whitley, D.
(2000). Matching horizon features using a messy ge-
netic algorithm. Computer Methods in Applied Me-
chanics and Engineering, 186:499–516.
Bierwirh, C., Mattfeld, D. C., and Kopfer, H. (1996). On
permutation representations for scheduling problems.
In Lecture Notes on Computer Science, volume 1141,
pages 310–318. Springer-Verlag, Berlin Heidelberg,
New York.
Brizuela, C. A. and Aceves, R. (2003). Experimental ge-
netic operators analysis for the multi-objective permu-
tation flowshop. In Lecture Notes on Computer Sci-
ence, volume 2632, pages 578–592. Springer-Verlag.
Chai, J. and Ma, S. (1998). An evolutionary framework for
stereo correspondence. In the 14th International Con-
ference on Pattern Recognition, pages 16–20, Bris-
bane, Australia.
Harris, C. and Stephens, M. (1988). a combined corner and
edge detector. In the Fourth Alvey Vision Conference,
pages 147–151, Manchester.
Jung, S. H. (2003). Queen-bee evolution for genetic algo-
rithm. Electronics Letters, 39(6):575–576.
Ruichek, Y., Issa, H., , and Postaire, J.-G. (2000). Genetic
approach for obstacle detection using linear stereo vi-
sion. In the IEEE Intelligent Vehicles Symposium,
Dearborn (MI), USA.
Schmid, C., Mohr, R., and Bauckhage, C. (2000). Evalua-
tion of interest point detectors. International Journal
of Computer Vision, 37(2):151–172.
Starkweather, T., McDaniel, S., and Mathias, K. (1991).
A comparison of genetic sequencing operators. In
Belew, R. and Booker, L., editors, the 4th Interna-
tional Conference on Genetic Algorithms, pages 69–
76, Morgan Kaufmann.
Yuan, X., Zhang, J., and Buckles, B. P. (2004). Evolution
strategies based image registration via feature match-
ing. Information Fusion, 5:269–282.
Zhang, B.-T. and Kim, J.-J. (2000). Comparison of selection
methods for evolutionary optimization. Evolutionary
Optimization, An International Journal on the Inter-
net, 2(1):55–70.
Zhang, Z., Deriche, R., Faugeras, O., and Luong, Q.-T.
(1994). A robust technique for matching two uncal-
ibrated images through the recovery of the unknown
epipolar geometry. Research report 2273, INRIA,
Sophia-Antipolis, France.
VISAPP 2008 - International Conference on Computer Vision Theory and Applications
644
