
LEVEL SET BRIAN SEGMENTATION WITH AGENT CLUSTERING
FOR INITIALISATION
Fast Level Set Based MRI Tissue Segmentation with Termite-Like Agent Clustering
for Parameter Initialization
David Feltell and Li Bai
School of Computer Science & IT, Nottingham University, Triumph Rd, Nottingham, UK
Keywords:
Level Set, Multi Agent Clustering, Image Segmentation.
Abstract:
This paper presents a novel 3D brain segmentation method based on level sets and bio-inspired methodologies.
Level set segmentation methods, although highly promising, require manual selection of seed positions and
thereshold parameters, along with manual reinitialisation to a new level set surface for each candidate region.
Here, the use of swarm intelligent mechanisms is used to provide all the statistical data and sample points
required, allowing automatic initialisation of multiple level set solvers. This is shown by segmentation of
white matter, grey matter and cerebro-spinal fluid in a simulated T1 MRI scan, followed by direct comparison
between a commercial level application - FMRIB’s FAST - and the ground truth anatomical model.
1 INTRODUCTION
The solution to initial value problems by level set
methods has transformed the study of evolving inter-
faces and, along with the surge in available processing
power, provides a unique and promising methodology
for three-dimensional image segmentation. However,
the initialization of surfaces and tuning of parameters
almost universally requires human intervention. By
using agent based clustering and segmentation algo-
rithms, we can hope to automate this process to a large
extent, thus no longer relying on expert knowledge or
requiring multiple tedious trials for an acceptable re-
sult.
The paper begins by reviewing the history of level
set methods, leading to the method currently used in
this work. Following this is a brief review of the
mechanisms of swarm intelligence, leading to previ-
ous attempts at data clustering and image segmenta-
tion using agent based principles. Next, the imple-
mentation details are given of the level set solver,
followed by the agent-based initialization algorithm.
Finally, preliminary results of the system are shown
along with a discussion of its merits and the project’s
future direction.
1.1 Level Set Method
Level set methods for image segmentation rely on an
evolving closed surface defined by a moving inter-
face, the front, which expands from a point out into
the image, fitting itself to the region it is released
within, and smoothing any noise. Among the advan-
tages of level set methods is the natural ability for
a single surface to seemingly split apart and merge
without losing its identity. To accomplish this we add
a further spatial dimension, φ, to our problem space,
then we can in effect have a single surface in (n+ 1)-
dimensional space, even though from observation of
n dimensions we see two or more spatially distinct
closed surfaces. This is the essence of the level set
method and allows for topological changes to be han-
dled naturally. This is easily visualised in the 2D case:
for example Figure 1, where two 2D surfaces are pro-
jected to a 3D volume, where it is shown they are
mathematically part of the same object. Numerically,
each point in space U includes a scalar φ that can vary
dynamically as time progresses, via a speed function
F. Functionally, by taking a slice at time t of all points
in U where φ = 0, we are taking the 0
th
level set of the
function φ.
If we initialise φ to the signed distance from an
initial closed surface, then we can visualise φ as the
height at a certain point, and all the points in space
with a height of 0 make up our surface. Thus, as well
as handling topological changes naturally, the level
set approach also has the advantage that the gradient
across the surface at any given point on the interface
can be found using ∇φ, meaning the local curvature
can be determined. A curvature term gives the signed
210
Feltell D. and Bai L. (2008).
LEVEL SET BRIAN SEGMENTATION WITH AGENT CLUSTERING FOR INITIALISATION - Fast Level Set Based MRI Tissue Segmentation with
Termite-Like Agent Clustering for Parameter Initialization.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 210-217
DOI: 10.5220/0001067702100217
Copyright
c
SciTePress
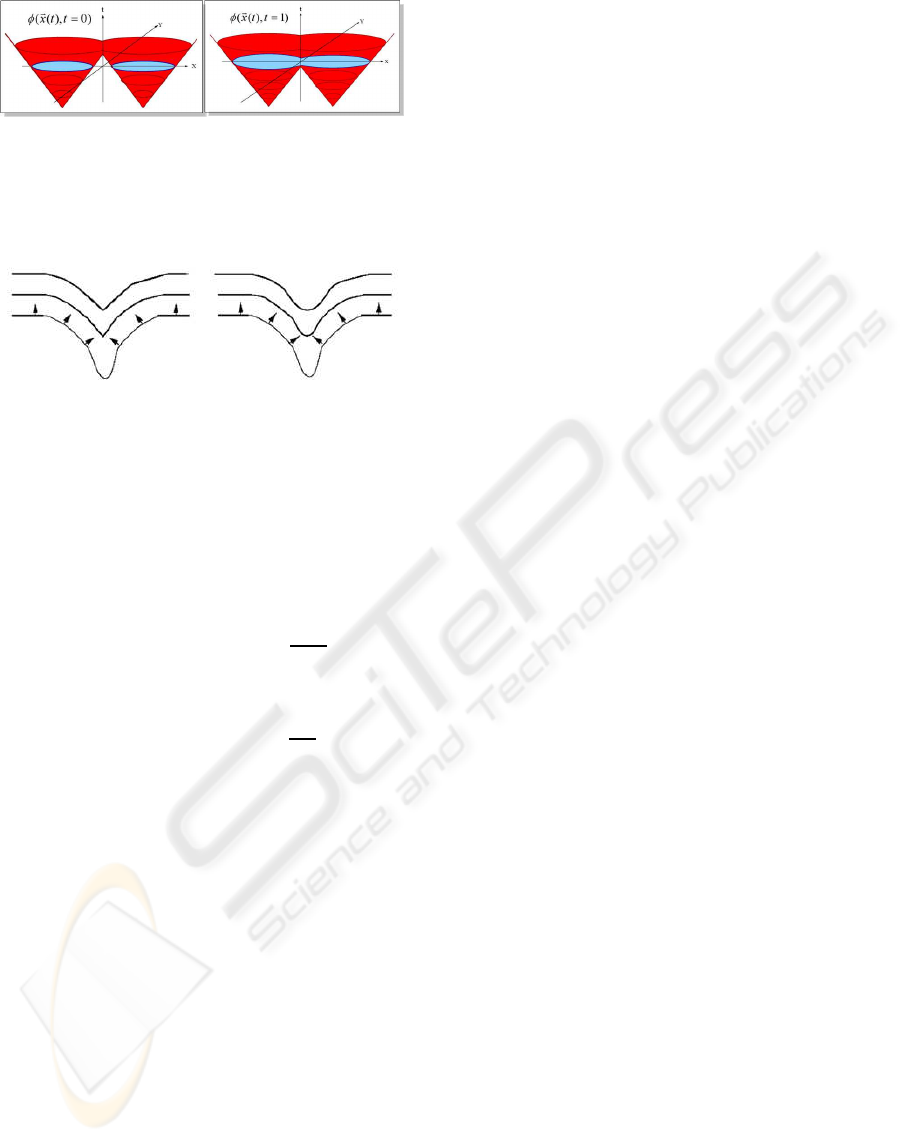
Figure 1: Illustration of level set function surface S (red)
and evolution of moving interface/zero level set (blue).
Topological change of two separate fronts (left) into a sin-
gle front (right) handled naturally via higher dimensional
function. After (Sethian, 1996).
Figure 2: Illustration of surface evolution without curva-
ture (left) and using a curvature term (right). After (Sethian,
1996).
’sharpness’ of the interface at a point, allowing for
a smoothing effect, overcoming noise and preventing
leaks. This is illustrated in Figure 2. The typical for-
mulation for the φ update function then becomes:
φ
t
= |∇φ|(αF + (1− α)∇•
∇φ
|∇φ|
) (1)
Where: φ
t
is the time derivative of φ; α controls
the the level of smoothness in the surface; F is the
data-dependent speed function; ∇ •
∇φ
|∇φ|
gives local
curvature at a given point.
Updating the values of φ at each point in space
requires a numerical technique to evolve the zero
level set from an initial specification. Naively then,
we could initialise a surface in space, create a suit-
able speed function F, and numerically integrate until
some condition is met and the surface extracted as all
points where φ = 0 (for example (Phillips, 1999)). An
advantage here is sub-cell accuracy - the φ values can
be interpolated to find points on a scale that is more
accurate than the discrete embedding it is operating
within. The first problem here is that the whole state-
space must be evaluated each iteration, rather than
just the surface. Secondly a small timestep is required
to prevent numerical instabilities. These two caveats
make the naive method undesirable for use outside of
specialist fields.
Algorithms have been developed to overcome
these issues by only performing updates on regions
near the surface, rather than integrating over the entire
state space. The most well-knownis the Narrow Band
approach (D. Adalsteinsson, 1995; Sethian, 1999),
where numerical integration is performed within a
band of points initialised around the surface, though
when the zero level set reaches the edge of this band,
the band must be reset. This dramatically increases
efficiency over the naive method, with little or no ef-
fect on accuracy. However, further optimization in
this vein has come in the form of the Sparse Field ap-
proach (Whitaker, 1998). With this method the nar-
row band is reduced to the smallest workable size and
the reinitialisation requirement is based on a purely
local update, rather than a global update of the entire
band. The reduction in accuracy is tolerable for most
applications and its implementation has allowed the
level set family of methods to achieve real-time per-
formance levels in complex 3D applications (for ex-
ample see (Lefohn et al., 2003; Lefohn et al., 2004)).
The Sparse Field approach has been taken even
further in the work of (Karl, 2005), and is the method
presented here. In this work the narrow band is low-
ered to simply two linked lists operating in a discrete
space, L
in
and L
out
, representing the inside and out-
side boundaries of the zero level set, respectively. No
attempt at sub-cell accuracy is made and most op-
erations use integer math. The values of φ are also
kept constant depending on their status and are up-
dated discretely. There are four such values repre-
senting inside, inside edge, outside edge, and outside
the volume, set so that a rough gradient can be found
at any point (for example: 3,1,−1,−3, respectively).
The level set algorithm approximation itself is imple-
mented in a two-pass process:
In the first pass elements in L
in
and L
out
are
checked using the speed function F. If an expan-
sion/contraction is provoked, the relevant list element
is switched to the opposing list. If an element finds
itself surrounded by cells with φ values of opposite
sign, it is removed from its list. This fact allows for
splits and merges in the topology.
The second phase involves an approximate Gaus-
sian smoothing term G, that is, taking the (weighted)
average of φ values from the area surrounding an el-
ement of L
in
or L
out
. Depending on the outcome of
G, expansion/contraction adjustments to the lists and
φ values are performed, similar to expansion via F.
This has the effect of smoothing away noise as well
as sharp protuberances in the surface.
Unlike previous algorithms, the process of expan-
sion via the speed function and contraction via curva-
ture are not intrinsically linked into the same update.
In this algorithm, F is run on L
in
and L
out
a number of
times. After this initial expansion phase a number of
G runs are similarly done on L
in
and L
out
. The ratio of
F to G runs determines the smoothness of the solution
(in an analogue to the α parameter in (1)).
LEVEL SET BRAIN SEGMENTATION WITH AGENT CLUSTERING FOR INITIALISATION - Fast Level Set Based
MRI Tissue Segmentation with Termite-Like Agent Clustering for Parameter Initialization
211

Further optimization can be made when the ratio
of F runs is much higher than G runs. Elements vis-
ited on the list that have reached a locally optimal po-
sition will remain in that position on the next run of
the speed function. Therefore they can be removed
from successive F runs on that pass.
As mentioned in their paper title, this method de-
parts almost entirely from the PDE based methods,
but maintains many of the advantages. Sub cell accu-
racy is not possible in this generic form, but it would
a simple addition to use this method as a prototype
generator for slower, but more accurate methods. The
inclusion of the φ embedding, albeit much simplified,
allows local topology to be determined at any point
to discrete-level accuracy. Thus, the Gaussian func-
tion has the information required to approximate the
’sharpness’ at a point, and to expand or contract as a
result (in a reasonable approximation to the effect of
curvature). The lack of the requirement to solve PDEs
holds several advantages. No timestep is required in
the update process - the algorithm is entirely discrete.
The method maintains the implicit ability of splits and
merges along the front, without relying on any gra-
dient calculation, eliminating the need for expensive
entropy-satisfying spatial derivative schemes.
Parameter selection for level set segmentation
generally requires at least some level of human in-
teraction. Specifically in the case of image segmen-
tation, initialization of a level set solver requires at
minimum seed locations, an ideal data value, and an
acceptable noise threshold to be preset. Also, differ-
ing data classes in the same problem space, for exam-
ple tissue types in medical scans, each require their
own level set surface with their own set of initializa-
tion parameters. Ideally the task of assigning seed
locations and calculating level set parameters would
be automated. These issues lend themselves well to
an agent based approach, which will be discussed in
the following section.
1.2 Multi-Agent Swarm Based
Algorithms
Agents are independent entities existing in some
environment - sensing, processing and modifying
the environment based on internal reasoning. For
multi-agent systems, the swarm intelligence and self-
organisation paradigms have become popularized in
many disciplines as an explanation for the apparent
mismatch in complexity of agent versus complexity
of task (Camazine et al., 2001), and as a unique engi-
neering metaphor (Bonabeau et al., 1999). Such sys-
tems of agents can focus on simple stimulus-response
functions using purely local stimuli with little or no
cognition or direct communication. Tasks are ac-
complished by exploiting the non-linearity inherent
in such a massively parallel system, rather than re-
lying on individual agent complexity. An agent mod-
ifying the environment at a particular location allows
another agent in that same location at a later time
to sense this new state and respond accordingly. In
this way the environment is used for indirect com-
munication, termed stigmergy. Stigmergy in bio-
logical systems is further enhanced by the complex-
ity of the environment, for example diffusion is uti-
lized in many biological processes to spread infor-
mation in the form of chemical gradients. Activa-
tion/attraction and inhibition/repulsion functions can
thus be designed to control agent interactions based
solely on local environmental state and any internal
state, relying on quantitative information reinforce-
ment and decay as well as qualitative signaling via the
environment to control the weighting between possi-
ble responses. A global-level task or structure may
then be many times more complex than an individual
can perceive or accomplish, yet through parallel ap-
plication of simple rules with indirect non-linear cou-
plings, we see the spontaneous emergence of a solu-
tion. Given this non-linearity, a tiny change in a pa-
rameter can result in a drastically different solution,
but (if the system is well formed) one that still con-
forms to a valid set of stable solutions - that is, the
system exhibits multistability. We have demonstrated
this engineering paradigm previously in modeling the
building behaviour of Macrotermes termites (Feltell
and Bai, 2004; Feltell et al., 2005). The use of an
attractive cement pheromone to coordinate soil clus-
tering behaviour has provided the inspiration for the
swarm-intelligent clustering mechanism used here.
Swarm based clustering algorithms have been de-
veloped to cluster sets of n-dimensional data, gen-
erally projected onto a 2D grid, by taking inspira-
tion from brood sorting and corpse clustering in ants
(Monmarche et al., 1999; Monmarche, 1999; Kanade
and Hall, 2003; Schockaert et al., 2004) as well as
building behaviour in termites (Vizine et al., 2005).
The approach does not need any prior knowledge of
the problem space or number of clusters, and as a
bonus gives a visual representation of the clusters on
the 2D grid.
Agent based approaches have also been devel-
oped to directly segment an image, again using in-
spiration from natural systems such as ant pheromone
trail networks (Ramos and Almeida, 2000), artificial
life (Liu et al., 1997; Liu and Tang, 1999; Bocchi
et al., 2005), social spiders (Bourjot et al., 2003), and
even termites, bloodhounds and children (Fledelius
and Mayoh, 2006). Here the agents interact directly
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
212

within the n-dimensions of the problem space, rather
than outside it. This distinction from swarm based
data clustering allows spatially localized regions to be
processed independently - possibly requiring only a
subset of the image space to be explored.
If the two perspectives can be combined, along
with other swarm-inspired mechanisms, we could
ideally form a localized data clustering algorithm,
whereby ideal seed locations and other parameters
would be found by agents within the image environ-
ment as they cluster similar voxels together.
Swarm intelligent systems emphasize robustness
and diversity over accuracy, and so find solutions to
complex problems that are ’good enough’, but which
require minimal agent capability in terms of both cog-
nition and interaction. One of the major lures of level
set algorithms is the tolerance for error, both in the
problem space and, within limits, the initialization
parameters. As long as ’good enough’ seed loca-
tion, ideal value and acceptable range parameters can
be determined, the solution to the level set equations
should be near-optimal in all but the hardest cases.
With this assumption in mind, well located seeds can
then be acceptable sample points used to approximate
the voxel mean and range within a class.
2 IMPLEMENTATION
2.1 Level Set Solver
The level set solver is derived from the model of Shi
& Karl (Karl, 2005). We define two sets of discretized
points, representing the inside and outside layers be-
tween which lies the level set surface. As well as up-
dating these sets as the surface expands or contracts
we must maintain the values of φ, such that φ = 3
inside the volume enclosed by the level set surface,
φ = 1 along the inside layer, φ = −1 along the outside
layer, and φ = −3 outside the volume (note: the sign
is reversed here compared to the original algorithm to
be more in line with other level set methodologies).
Finally, we must ensure that no orphaned points exist.
That is, inside points must always have at least one
outside point adjacent to them, and similarly outside
points must always have at least one inside point ad-
jacent. The update routine for both inside and outside
layers is similar, though varies in parameter value.
Following is a summary of the general update routine:
Let:
• Current time step be t.
• Two sets of points defining our two discretized
layers be S
i
and S
j
.
• φ constants in space be: b
i
along S
i
; b
j
along S
j
; a
i
in the volume beyond S
i
; a
j
in the volume beyond
S
j
.
• Three-dimensional cartesian point vector, p =
(pi+ p j + pk).
• Neighbourhood of p, N(p) = {(pi + i + pj +
pk),(pi − i + pj + pk),(pi + pj + j + pk),(pi +
pj − j + pk), (pi + p j + pk + k),(pi+ pj + pk −
k)}.
• Context dependent speed function, F(p) : ℜ −→
ℜ, with condition f
i
required for update to occur.
Here and throughout this work it is assumed voxel
intensity value is in [0,1] ∈ ℜ.
Then, for each p ∈ S
i
, iff F(p) =⇒ f
i
:
1. Increment time step, t.
t ←− t + 1 (2)
2. p is added to S
j
S
j
←− {S
j
∪ {p}} (3)
3. φ at point p is set to constant, b
j
.
φ(p,t) ←− b
j
(4)
4. p is removed from S
i
, whilst all relevant neigh-
bours of p are added to S
i
.
S
i
←− {S
i
− {p}} ∪ {∀r ∈ N(p) | φ(r,t) = a
i
}
(5)
5. φ at all relevant neighbours of point p is set to
constant, b
i
.
φ({r | φ(r,t) = a
i
},t) ←− b
i
,∀r ∈ N(p) (6)
6. All points in S
i
that are in not neighboured by a
point in S
j
are orphaned points and must be re-
moved.
S
i
←− S
i
− {∀r ∈ S
i
| N(r) ∩ S
j
=
/
0} (7)
7. φ at all orphaned points in S
i
is set to constant, a
j
.
φ({r | N(r) ∩ S
j
=
/
0},t) ←− a
j
,∀r ∈ S
i
(8)
In practice, all these steps can be combined into a sin-
gle update loop. The update loop is performedon both
the inside and outside layers. Let our inside surface
be S
in
and the outside surface be S
out
, then: a
in
= 3,
a
out
= −3, b
in
= 1, b
out
= −1, f
in
⇔ (F(p) > 0),
f
out
⇔ (F(p) < 0). The steps (3)..(6) deal with the
surface evolution. The steps (7) and (8) handle shock
propagation, that is, they remove points as the curve
crosses over itself to maintain an unambiguous closed
surface.
LEVEL SET BRAIN SEGMENTATION WITH AGENT CLUSTERING FOR INITIALISATION - Fast Level Set Based
MRI Tissue Segmentation with Termite-Like Agent Clustering for Parameter Initialization
213

The algorithm is then:
Let:
• D(p) be the problem specific data term, specifi-
cally the voxel gray level value at point p.
• J(p) be the problem specific speed function,
specifically: J(p) = 1 for |(D(p) − T)| < ε and
J(p) = −1 for |(D(p) − T)| > ε, where: T is the
ideal data value; ε is the acceptable error.
• G(p) be a Gaussian smoothing approximation,
specifically G(p) =
φ(p,t) +
∑
r∈N(p)
φ(r,t)
/7.
With the condition G(p) = 0 iff |G(p)| < 1, tighter
areas can be explored, as smoother curves will
neither contract nor expand. This produces more
accurate solutions in most cases, but tends to give
less smooth surfaces.
• t
J
be the number of speed runs to perform; t
G
be
the number of Gaussian smoothing runs to per-
form.
Then:
A Initialize S
in
and S
out
to small surface(s) about
given seed locations.
B Perform (2)..(8) with: i = out; j = in; F(p) =
J(p).
C Perform (2)..(8) with i= in; j = out; F(p) = J(p).
D Repeat from (B) while t < t
J
.
E Perform (2)..(8) with: i = out; j = in; F(p) =
G(p).
F Perform (2)..(8) with i = in; j = out; F(p) =
G(p).
G Repeat from (E) while t < t
J
+ t
G
.
H Set t = 0. Repeat from (B) for t
S
iterations.
2.2 Swarm based Parameter
Initialization
The parameter specification for the level set solver as
well as the number of tissue typesrequiring individual
segmentation is beyond the capability of a level set al-
gorithm alone. Instead, in this work we show how we
can use a collection of agents following similar rules
to previous agent clustering algorithms, but with the
agents embodied within the image space itself, find-
ing good seed locations as well as performing minor
preprocessing functions.
Agents have real-valued position and direction,
using simple nearest neighbour approximation when
sensing the underlying discrete image voxels. They
wander the image in an initially random direction,
however as they move they lay a quantity of attrac-
tive ’pheromone’ in visited voxels. The quantity of
pheromone is proportional to the divergence of voxel
intensity |∇
2
· D| at their current location. A high di-
vergence value often indicates a heterogeneity in the
image - specifically an interface between two regions.
Here, pheromone deposition further has the restric-
tion D > 0.1 to avoid segmenting irrelevant black re-
gions. Pheromone diffuses through the image lattice,
with the edge of the lattice and near-black (D < 0.1)
voxels acting as sinks. The addition of a positive re-
inforcement pheromone mechanism allows the agents
to use gradient following behaviour in coordinating
toward promising seed locations. As pheromone dif-
fuses away it causes that area to receive less attention,
ultimately meaning less suitable seed locations are
abandoned. This reflects in many ways the clustering
behaviour of termites in nest construction (Bruinsma,
1979), where heterogeneities stimulate deposits of
soil laden with a cement pheromone, which in turn
attracts other termite builders to the site.
This clustering behaviour, in the termite case, cre-
ates regular spaced piles or pillars of soil. Piles close
to one-another thus compete for the termites atten-
tions, eventually resulting in regularly spaced pillars.
In this work, as with the termite paradigm, this mech-
anism means seed locations tend not to become too
localised. The combined effect ultimately increases
the agents’ sampling efficiency within large images.
The generalised update routine for an individual
agent is given as follows:
Let:
• t be the current time step of an agent.
• Real-valued point vector, p be the current position
of an agent
• Scalar pheromone field, ρ(p) be the pheromone
concentration at location p.
• Speed, s be a scalar speed value for the agent.
• Vector v be the continuous direction vector of the
agent.
Then:
1. Increment time step, t.
t ←− t + 1 (9)
2. Lay pheromone proportional to the logarithm of
the absolute divergence. Logarithmicscale is used
here to avoid floating point accuracy overflow.
log(ρ(p)) ←− log
ρ(p) + |∇
2
D(p)|
(10)
3. Calculate new direction vector from weighted gra-
dient of logρ.
v ←− v+ α(v− ∇logρ) (11)
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
214
4. Calculate speed scalar based on local density of ρ.
s ←− 1+
β
1+ log(ρ(p))
(12)
5. Calculate updated position based on normalized
direction vector and speed value.
p ←− p+
v
||v||
· s (13)
Where: α is the weighting of pheromonegradients
in movement; β controls the maximum speed of an
agent.
The full agent algorithm is then:
A Initialize set of n agents, M, each with random
position within the image and random direction.
B Perform (9)..(13) for each m ∈ M.
C Diffuse pheromone: ∂
t
ρ = d · ∇
2
ρ
D Repeat from (B) while t < t
A
time steps.
E Select the top C voxel locations with highest
pheromone level ρ, to get full seed list, Q.
F Run k-means clustering on Q, classifying into
prespecified number of sets, Q
i
, where i =
{1,2,..,q}.
G Within each Q
i
calculate mean µ(Q
i
) and standard
deviation σ(Q
i
), givingideal data value T = µ(Q
i
)
and error threshold ε = σ(Q
i
) + k, where k is a
constant, within a class.
Once T and ε have been calculated for a seed set
Q
i
, the level set routine can be run, initialising L
in
and L
out
surfaces as pseudo-spheres about each seed
location in Q
i
. The agent algorithm need only be run
once, then the level set algorithm run once for each
seed set.
Additionally, in practice the diffusion step can
be moved into a separate thread to take advantage
of modern day multiple core processors. With the
simplest distributed setup, with little control on syn-
chronization or load balancing, the algorithm can still
function correctly - an implicit advantage of the ro-
bust swarm metaphor.
The image is finally extracted as all voxel posi-
tions where φ > 0, thus all points lying on and within
L
in
.
3 RESULTS
The dataset used comes from BrainWeb’s online MRI
simulator (McConnell BIC, 2007) with default pa-
rameters: modality T1, slice thickness 1mm, noise
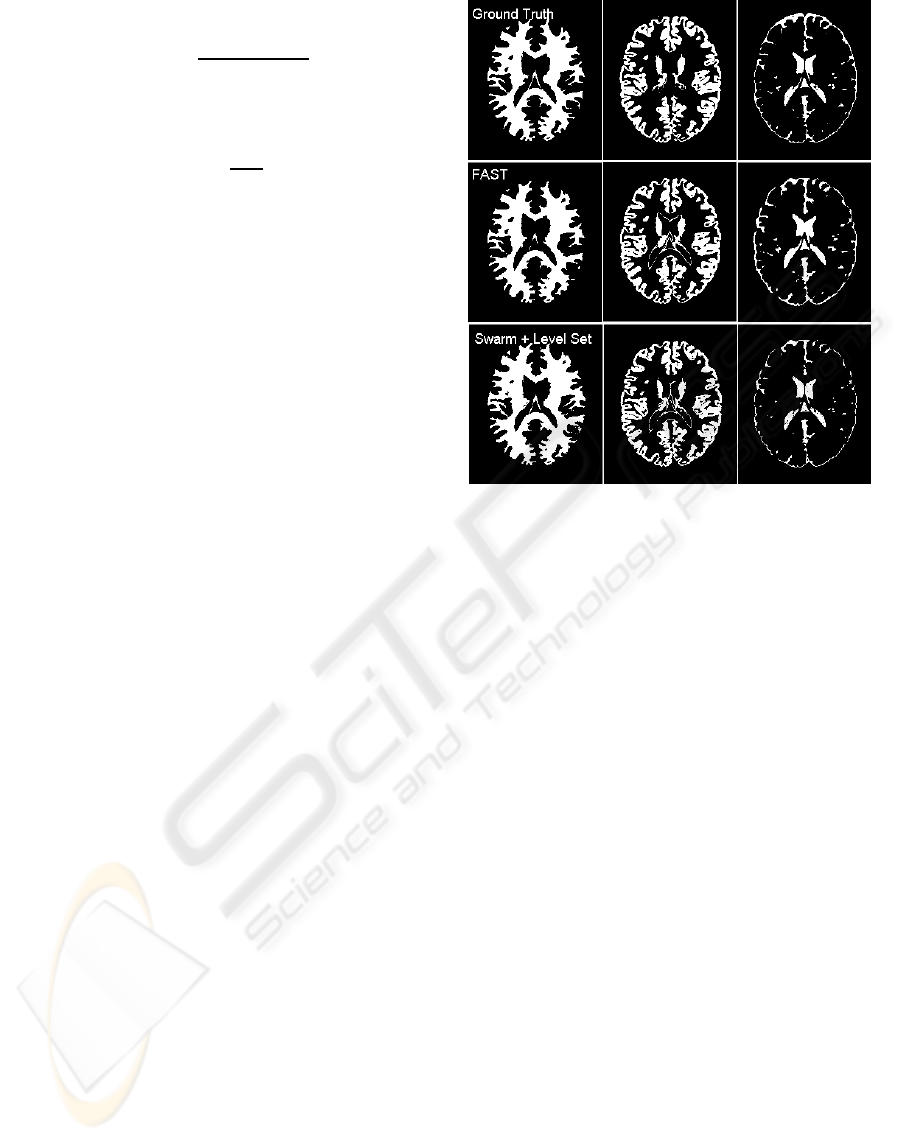
Figure 3: Comparison of ground truth to FAST and the
swarm initialised level set solution presented here. Images
used in both FAST and the present work have been prepro-
cessed by BET. From left to right: white matter; gray mat-
ter; cerebro-spinal fluid.
3% and non-uniform intensity 20%. For comparison
with ground truth and FAST (Zhang et al., 2001) the
number of classes is set q = 3. The various parame-
ters, unless otherwise stated, are set as follows:
α = 2;β = 2.5;d = 0.015;k = 0.02;t
S
= 10;t
A
=
3000;t
J
= 30;t
G
= 3
The parameter values above for α,β,d and k, were
found in part using a real-valued genetic algorithm,
using the similarity to the ground truth images, mi-
nus the similarity between one another, as a fitness
function. This process has proven useful so far, yet
is only partially utilised here and remains a direction
for future work. Other values are chosen intuitively to
balance execution time and accuracy.
From Figure 3 it can be seen how the approach,
even in this early stage, can compete with popular so-
lutions such as FMRIB’s FAST, which uses a method
based on Markov random fields. In both solutions,
preprocessing has been performed using FMRIB’s
popular BET (Smith, 2002) to skull strip and nor-
malise the image voxels.
Quantitative measurement on accuracy is approx-
imately possible given the hand segmented ground
truth images available. In the above case, FAST
achieves approximately 92%, 85% and 57% accu-
racy, whereas the swarm initialised level set algorithm
LEVEL SET BRAIN SEGMENTATION WITH AGENT CLUSTERING FOR INITIALISATION - Fast Level Set Based
MRI Tissue Segmentation with Termite-Like Agent Clustering for Parameter Initialization
215

achieves 86%, 83% and 55%, for white matter, grey
matter and cerebro spinal fluid, respectively. FAST
remains superior, but the small score difference cer-
tainly indicates the presented approach is well worth
further investigation.
In terms of execution time, from the point of file
loading to final file output, FAST outdoes the perfor-
mance of the presented work by just over double. The
above solution was found by the swarm initialised
level set algorithm in approximately 17 mins, whereas
FAST finished in approximately 8 mins. It is worth
noting that the swarm approach presented is still in
early development, and as discussed in the next sec-
tion, has the advantage of naturally supporting a dis-
tributed implementation.
4 DISCUSSION AND FUTURE
WORK
The level set solver is an approximate discrete solu-
tion to a continuous problem. Although the algorithm
used in this work is particularly fast, it simply does
not have the power of a narrow band or even sparse
field approach. These methods not only reflect the
naive approach more directly, but also have sub-cell
accuracy, where actual surface points can be interpo-
lated within image voxels using the real-valued gradi-
ent of φ. They can also vary in speed along different
areas of the surface, allowing for a more global curva-
ture force effect. The discretisation of φ values to sim-
ply the set {−3,−1, 1, 3} means the curvature term is
much more rigid, and does not distinguish on a lo-
cal level between corners of differing sharpness. Ide-
ally the current level set solver approximation would
work as a rapid prototype for a more accurate narrow
band based method. This remains a direction for fu-
ture work.
One benefit of using a swarm based system is in
the robustness to a noisy environment. In this regard
it should be possible to include an atlas mechanism,
using only the most minimal registration with the can-
didate image, and restrict agent interaction within or
near the atlas template for a given region. This would
give the agents’ environment a significant amount of
extra information, which can be used to focus agent
efforts - increasing efficiency and allowing for seg-
mentation of more difficult (too smooth/too noisy) re-
gions.
The preprocessing by BET could be removed for
future versions. BET simplifies segmentation signif-
icantly, and prevents areas such as the skull being
segmented as part of brain matter. However, pre-
liminary results of segmentation without BET pre-
Figure 4: Solution found using current algorithm without
BET preprocessing. All parameters are set similar - the
number of classes remaining at 3. From left to right: white
matter, gray matter, cerebro-spinal fluid.
processing are promising in their own right, though
show several mismatched regions (see Figure 4), and
it is likely that with more classes and ground truth
samples, along with evolutionary algorithm parameter
tuning, the method can be applied to multiple modal-
ities and varying image spaces without the need for
any preprocessing.
Another benefit of the multi-agent paradigm is its
naturally distributed nature. As has been eluded to,
the diffusion of pheromone can already be ported to
another processing unit to greatly increase efficiency.
A further extension could see the image space and/or
agents split to run parallel. Certainly each region’s
level set solver could in future run on separate pro-
cesses to dramatically decrease execution time.
REFERENCES
Bocchi, L., Ballerini, L., and Hssler, S. (2005). A new evo-
lutionary algorithm for image segmentation. Applica-
tions on Evolutionary Computing, pages 264–273.
Bonabeau, E., Dorigo, M., and Theraulaz, G. (1999).
Swarm Intelligence: From Natural to Artificial Sys-
tems. Oxford University Press.
Bourjot, C., Chevrier, V., and Thomas, V. (2003). A new
swarm mechanism based on social spiders colonies:
From web weaving to region detection. Web Intelli-
gence and Agent System, 1(1):13–32.
Bruinsma, O. (1979). An Analysis of Building Behaviour of
the Termite Macrotermes Subhyalinus (Rambur). PhD
thesis, Landbouwhogeschool, Wageningen.
Camazine, S., Deneubourg, J.-L., Franks, N., Sneyd, J.,
Theraulaz, G., and Bonabeau, E. (2001). Self-
Organization in Biological Systems. Princeton Uni-
versity Press.
D. Adalsteinsson, J. S. (1995). A fast level set method for
propagating interfaces. J. Comp. Phys., 118:269–277.
Feltell, D. and Bai, L. (2004). Swarm robotics for con-
struction. In 24th SGAI International Conference on
Innovative Techniques and Applications of Artificial
Intelligence, Cambridge, UK.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
216

Feltell, D., Bai, L., and Soar, R. (2005). Bio-inspired emer-
gent construction. In IEEE 2nd Intl. Swarm Intelli-
gence Symposium, Pasadena, CA, USA.
Fledelius, W. and Mayoh, B. (2006). A swarm based ap-
proach to medical image analysis. In 24th IASTED In-
ternational Conference on Artificial Intelligence and
Applications (AIA 2006), pages 150–155, Anaheim,
CA, USA. ACTA Press.
Kanade, P. and Hall, L. (2003). Fuzzy ants as a cluster-
ing concept. In 22nd International Conference of the
North American Fuzzy Information Processing Soci-
ety (NAFIPS), pages 227–232.
Karl, Y. S. W. (2005). A fast level set method without solv-
ing pdes. In IEEE International Conference on Acous-
tics, Speech, and Signal Processing (ICASSP 2005),
volume 2, pages 97–100.
Lefohn, A. E., Cates, J. E., and Whitaker, R. T. (2003). In-
teractive, gpu-based level sets for 3d segmentation. In
Medical Image Computing and Computer-Assisted In-
tervention (MICCAI 2003), pages 564–572.
Lefohn, A. E., Kniss, J. M., Hansen, C. D., and Whitaker,
R. T. (2004). A streaming narrow-band algorithm: In-
teractive computation and visualization of level sets.
IEEE Transactions on Visualization and Computer
Graphics, 10:422–433.
Liu, J. and Tang, Y. (1999). Adaptive image segmentation
with distributed behavior-based agents. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
21:544–551.
Liu, J., Tang, Y., and Cao, Y. (1997). An evolutionary
autonomous agents approach to image feature extrac-
tion. IEEE Transactions on Evolutionary Computa-
tion, 1:141–158.
McConnell BIC (2007). Brainweb: Simulated
brain database, Montreal Neurological Institute.
http://www.bic.mni.mcgill.ca/brainweb/.
Monmarche, N. (1999). On data clustering with artificial
ants. In Freitas, A. A., editor, Data Mining with Evo-
lutionary Algorithms: Research Directions, pages 23–
26, Orlando, Florida. AAAI Press.
Monmarche, N., Slimane, M., and Venturini, G. (1999). On
improving clustering in numerical databases with arti-
ficial ants. In ECAL ’99: Proceedings of the 5th Euro-
pean Conference on Advances in Artificial Life, pages
626–635, London, UK. Springer-Verlag.
Phillips, C. L. (1999). The level set method. The MIT Un-
dergraduate Journal of Mathematics, 1:155–164.
Ramos, V. and Almeida, F. (2000). Artificial ant colonies in
digital image habitats - a mass behaviour effect study
on pattern recognition. In 2nd Int. Workshop on Ant
Algorithms (ANTS 2000), pages 113–116, Brussels,
Belgium.
Schockaert, S., Cock, M. D., Cornelis, C., and Kerre, E. E.
(2004). Fuzzy ant based clustering. In International
Workshop on Ant Colony Optimization and Swarm
Intelligence (ANTS 2004), pages 342–349, Brussels,
Belguim.
Sethian, J. (1996). Level set methods: An act of vio-
lence - evolving interfaces in geometry, fluid mechan-
ics, computer vision and materials sciences. American
Scientist.
Sethian, J. (1999). Level Set Methods and Fast Marching
Methods. Cambridge University Press.
Smith, S. (2002). Fast robust automated brain extraction.
Human Brain Mapping, 17(3):143–155.
Vizine, A., de Castro, L., Hruschka, E., and Gudwin, R.
(2005). Towards improving clustering ants: An adap-
tive ant clustering algorithm. Informatica, 29(2):143–
154.
Whitaker, R. (1998). A level-set approach to 3d reconstruc-
tion from range data. International Journal of Com-
puter Vision, 29(3):203–231.
Zhang, Y., Brady, M., and Smith, S. (2001). Segmentation
of brain mr images through a hidden markov random
field model and the expectation maximization algo-
rithm. IEEE Trans. on Medical Imaging, 20(1):45–57.
LEVEL SET BRAIN SEGMENTATION WITH AGENT CLUSTERING FOR INITIALISATION - Fast Level Set Based
MRI Tissue Segmentation with Termite-Like Agent Clustering for Parameter Initialization
217
