
MEDICAL KNOWLEDGE REPRESENTATION WITHIN
HEARTFAID PLATFORM
Dragan Gamberger, Marin Prcela, Alan Jović, Tomislav Šmuc
Rudjer Boskovic Institute, Bijenicka 54, Zagreb, Croatia
Ganfranco Parati, Mariaconsuelo Valentini
Cardiology II, S.Luca Hospital, Istituto Auxologico Italiano, Milano, Italy
Kalina Kawecka-Jaszcz, Katarzyna Styczkiewicz, Andrzej Kononowicz
I Cardiac Department, Jagiellonian University Medical College, Cracow, Poland
Antonio Candelieri, Domenico Conforti, Rosita Guido
Department of Electronics, Informatics and Systems, University of Calabria, Rende (Cosenza), Italy
Keywords: Heart failure, knowledge representation, ontology, production rules.
Abstract: The paper presents the results of the development of the knowledge base for the HEARTFAID platform.
The means and methods used to collect, systematize and formalize medical knowledge, as well as to test the
developed knowledge representation are described. The descriptive part of the knowledge base is realized as
an ontology which conceptualizes the heart failure medical domain. The procedural part of the knowledge
base is realized through sets of production rules. The procedural knowledge covers the tasks of heart failure
diagnosis, severity assessment, treatment process, medication prescription and dosage, medication
contraindications, prognosis estimation, and acute decompensation detection. Finally, medical plans are
used to present medical actionable knowledge. Currently they are used only to systemize procedural
knowledge development process but they present a challenge for the future work in the field of medical
knowledge representation.
1 INTRODUCTION
Medicine is the field characterized by the enormous
amount of existing expert knowledge and at the
same time there is a need for constant and reliable
decision making. This is an ideal scenario for
building and using automated knowledge based
decision systems. Building an effective knowledge
base is a challenge relevant not only for the
Heartfaid platform, but for all artificial intelligence
applications as well. It is also known as a hard
problem with possibly many different solutions
among which none can be selected as ideal or
optimal for all situations.
The knowledge representation is actually a very
lively research field, especially in medical
application. Heartfaid platform is a good example of
a real medical environment for which automated
intelligent decision making is necessary. In our work
we first tried to test different knowledge
representation options and then to select and use
modern and most appropriate technology for solving
concrete decision making problems.
2 MEDICAL MOTIVATION
Heart failure is increasingly frequent in western
world, carrying a high mortality rate and being
responsible for a consistent increase in healthcare
costs related to the multiple therapeutic interventions
307
Gamberger D., Prcela M., Jovi
´
c A., Šmuc T., Parati G., Valentini M., Styczkiewicz K., Kawecka-Jaszcz K., Kononowicz A., Candelieri A., Conforti D. and
Guido R. (2008).
MEDICAL KNOWLEDGE REPRESENTATION WITHIN HEARTFAID PLATFORM.
In Proceedings of the First International Conference on Health Informatics, pages 307-314
Copyright
c
SciTePress

and the high frequency of hospital admissions
required by these patients. There is therefore an
increasing need for a better care, that might be
provided not only by highly specialized centers, but
also by small hospitals and by field cardiologists, a
need that has to be matched with a policy of cost
containment. Progress in technology may offer an
important support to make this possible, allowing
adequate knowledge to be made available to all
health care providers in this field. It might also offer
new methods for regular and accurate collection of
biological signals in patients living in their home
environment, making use of sensors, either
traditional or wearable, able to provide a continuous
monitoring also through telemedicine facilities.
More recent progress might further offer an
advanced platform of services for the automated
integration between the signals collected both at
home and in the clinic environment on one side, and
the available state-of-the-art knowledge on the other
side, providing an artificial intelligence support to
clinical decision. The proper adoption of these tools
might help improving the daily care of chronic heart
failure patients, through a prompt titration of
treatment in response to early detection of even
minor changes in clinical conditions, as well as
through a reduction of diagnostic and therapeutic
errors, by reinforcing the implementation of the
most advanced recommendations provided by
international clinical guidelines. Such an approach
might also help improving the cost/effectiveness of
heart failure care facilitating the implementation of a
disease management approach, in which therapy,
education and follow-up are tailored for each patient
by a multidisciplinary team constantly supported by
an advanced platform of computerized services
guiding the clinical decision through a continuous
update of patient’s clinical conditions allowed by
advanced wireless telecommunication technology.
The Heartfaid project is aimed at developing such a
tool and at testing its feasibility and usefulness in the
management of chronic heart failure patients,
focussing in particular on those with advancing age.
3 RELATION WITH DECISION
SUPPORT SUBSYSTEM
Decision support subsystem (DSS) is the part of the
platform responsible for its intelligent behaviour and
the knowledge base (KB) is the representation of the
medical knowledge necessary for the DSS
operability. In order that this knowledge can be used
by the DSS, it must be presented in a formally sound
way. The task of building the knowledge base
consists of collecting the relevant medical
knowledge, its systematization, and technical
formalization.
User services are not supposed to directly access
the knowledge available in the KB. They can only
ask for the assistance of the DSS, which can then
decide to use the KB for its decision making
process. It means that during normal platform
operation, the KB, with exception of DSS, is isolated
from other platform parts. In contrast to that, during
the platform development the KB is perhaps the
most relevant integrative part between medical and
technical partners. Building it presents the challenge
of transferring all aspects of relevant medical
knowledge into the platform. The success in this
work significantly determines the overall
performance of the system.
On the other side, DSS completely determines
the requirements set on formal aspects of the
knowledge representation task. The final goal is
effective interplay between the DSS and the KB.
Already in the project definition it has been
determined that the ontological form of knowledge
representation is the most appropriate for the
concrete task. In the DSS development phase the
semantic web OWL ontology form with integrated
rules in the SWRL form have been selected as
optimal solution. The main reason for these
decisions is that only relative simple representation
forms without explicit time component and with
deterministic logic can be effectively handled by
available open source interpreters and reasoning
systems.
In our work we have also experimented with
some other systems, especially those specifically
designed for medical applications and guideline
modelling. We have worked with Arden syntax,
GLIF, Asbru, and PROforma.
Arden syntax (Hripcssak et al, 1993) is a rule-
based system adopted in the year 1992 and it is now
part of HL7 standard. The main idea of the Arden
syntax is to add as much as possible human-readable
information to machine-readable rules. Each rule is
stored in a single file and is called a Medical Logic
Module (MLM). The drawback of Arden syntax is
that it does not refer to any kind of domain
description (ontology). Due to this, the system needs
to interact directly with a clinical database in order
to provide alerts and reminders, which strongly
hiders knowledge sharing. Also, the execution
components are not freely available (mostly because
of Arden’s institution dependency). However, unlike
HEALTHINF 2008 - International Conference on Health Informatics
308

the vast majority of other systems, it has found a
practical usage in real clinical environments.
GLIF (Peleg et al, 2000) provides a framework
for developing medical guidelines that are both
easily understandable by humans (medical experts)
and interpretable by machines. Each GLIF guideline
is modelled in the form of a flowchart (directed
graph). GLIF is suitable for describing logic
sequence of actions. Within the HEARTFAID
platform GLIF may be used to represent the logical
flow of actions, e.g. sequence of tests performed for
diagnosing disease or prescribing therapy but the
problem is that there exists only commercial
execution engine (Glee).
Asbru (Shahar et al, 1998) is a guideline
modelling tool which focuses on representing
medical plans. It is highly aware of the time
dimension in the medical procedures and actions. A
plan in Asbru is a set of actions that are performed
when certain preconditions hold. Each plan is
decomposed into more sub-plans that are performed
sequentially, concurrent (parallel execution) or
cyclical. Within the HEARTFAID platform, Asbru
can be used in situations where actions are taken in a
predefined order, e.g. to describe the procedure at
the baseline evaluation or additional patient visits to
the clinics. However, there are no freely available
execution engines that may be integrated into
HEARTFAID platform.
PROforma (Sutton et al, 2003) is a knowledge
composition language that aims to assist patient care
through active decision support and workflow
management. Similar to the GLIF model, it
represents also guidelines as a directed graph in
which nodes represent instances from the PROforma
task ontology. PROforma contains a number of tools
for developing guidelines. A major focus point is on
guideline safety by defining additional safety-related
operators such as integrity and safety constraints.
Considering the execution engines, Arezzo is a
commercial version of PROforma, while Tallis is a
version available for educational and research
purposes (under license agreement).
4 DESCRIPTIVE HEART
FAILURE KNOWLEDGE
The first step in the development of the knowledge
base for the Heartfaid platform has been
development of the heart failure (HF) ontology. It
presents the formalized description of concepts for
the whole heart failure domain. It includes basic HF
concepts, properties that characterize patients, all
relevant diagnostic examinations and tests, and
treatment procedures. The ontology also includes
other cardiovascular system related concepts as well
as concepts related to other organs when they are
connected with HF. The information presented in the
ontology has been obtained by human interpretation
of guidelines for congestive and acute heart failure
(http://www.escardio.org/knowledge/guidelines/),
Heartfaid reports, as well as from other medical
knowledge sources, including, but not limited to
UMLS (Unified Medical Language System), Mayo
clinic web site and Open Clinical web site.
In its current form the ontology presents the
detailed taxonomic overview of the HF domain with
around 200 classes describing HF related concepts.
Examples are "Cardiac_hypertrophy", "Blood_
pressure_signs" or "Heart_murmurs". These
concepts are interconnected with super-class and
sub-class properties into a hierarchical tree-like
structure. At the basic level there are five relavant
super-classes: "HF_concept", "Patient_characte-
ristic", "Patients", "Testing", and "Treatment".
Figure 1 presents the Protégé tool displaying these
five super-classes with some of their most relevant
sub-classes.
Individuals or instances are members of the
classes and typically present exhaustive list of
concrete concepts relevant for the class. For
example, the "Cardiac_hypertrophy" class has
following six instances: "Cardiomegaly",
"Combined ventricular hypertrophy", "Left_atrial_
hypertrophy", "Left_ventricular-hypertrophy",
"Right_atrial_hypertrophy", and "Right_ventricular_
hypertrophy". The ontology includes more than
2000 individuals. When possible, classes are
specified with their CUI number (Concept Unique
Identifier according to UMLS) and with a list of
synonyms. For example, for the class
"Heart_diseases" its CUI is C0018799 and its
synonyms are "Disorder_of_heart", "Cardiac_
diseases", "Cardiopathy".
Finally, the ontology contains properties that
connect individuals in different classes. These
properties are relevant because they enable
introduction of relations among concepts. For
example, individual "Valvular_heart_disease" from
the class "Heart_valve_diseases" is indicated by the
individual "Dyspnea" from the class of
"Signs_and_symptoms". Or that "Hyperkalemia"
from the class "Potassium_disorder" may be caused
by medications like "Potassium_sparing_diuretics"
or "Spironolactone". The names of these properties
are "Indicated" and "MayBeCausedByMedication".
MEDICAL KNOWLEDGE REPRESENTATION WITHIN HEARTFAID PLATFORM
309
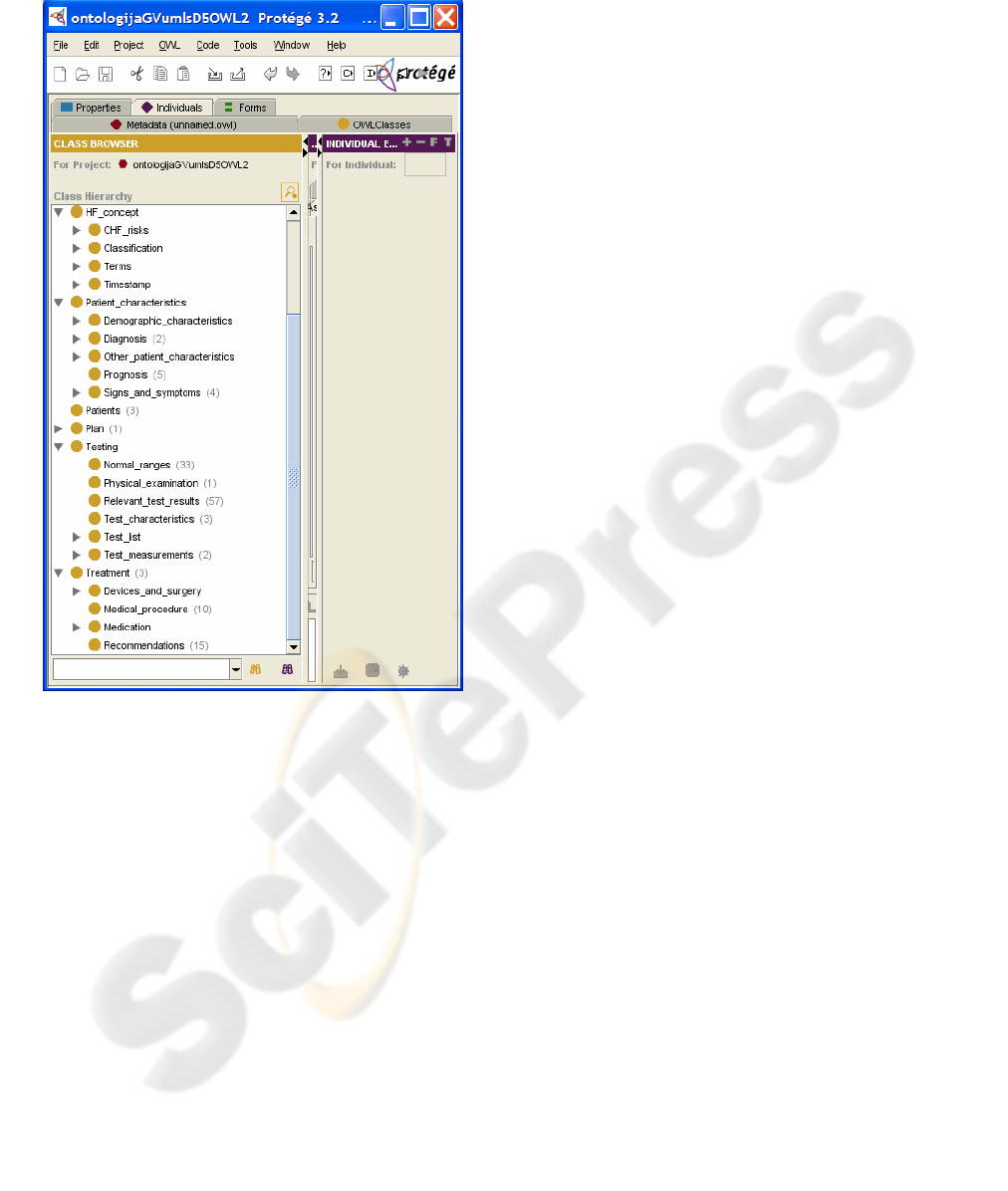
The HF ontology includes definitions of more than
100 properties.
Figure 1: Protégé tool used to display a part of the HF
ontology.
The ontology presents descriptive domain
knowledge. This knowledge is of two types. The
first is defined by the generality relations among
instances and classes, as well as by the generality
relations among sub-classes and super-classes. In
this way for each concept presented by some
instance there is a series of is-a relation. For
example, it means that "Cardiomegaly" is-a
"Cardiac_hypertrophy" while "Cardiac_
hypertrophy" is-a "Heart_disease". The second type
of descriptive knowledge contained in the ontology
comes from properties that define relations between
classes, such as "Indicated" or "MayBe
CausedByMedication", mentioned before.
Descriptive HF ontology is publicly available in
the web form from the project web site at
http://www.heartfaid.org/links.php.
5 PROCEDURAL KNOWLEDGE
The HF ontology presents the detailed taxonomic
overview of complete heart failure domain including
relevant relations among concepts. It represents
descriptive knowledge about the domain. Platform
should also be able to perform some actions,
typically in the form of suggestions for patients and
medical personnel. The knowledge representing
sufficient and necessary conditions that some actions
can be done is the so called procedural knowledge.
Descriptive and procedural knowledge together
present the knowledge base of the HF platform.
5.1 Production Rules
Production rules in a form "IF some condition is true
THEN make some action" are a widely used
approach for the presentation of procedural
knowledge. Their advantages are natural
interpretation by humans and modularity during
construction. It is also relevant that production rules
are also a formal way of presenting knowledge and
in this way a good starting point for practical
realization of the decision support system. For the
integration of descriptive and procedural knowledge
it is important that production rules can use only the
concepts defined in the HF ontology.
At the knowledge presentation level it is very
important that production rules can be easily
understood and corrected by medical experts. In this
way the major advantage of presenting procedural
knowledge in the form of production rules is that
they present formal enough way to present
knowledge that can be used by the platform and that
at the same time medical experts can easily control
the expected performance of the platform. The
correction of rules or adding them should only be
performed by the authorized medical personnel.
The HF procedural knowledge has been divided
into 10 functional subtasks in order to enable easier
human control of the completeness and consistency
conditions. They are:
1. HF diagnosis
2. Alternative or additional diagnosis
3. Heart failure severity assessment – specifying
NYHA class for patient, which is required for
general patient treatment approach
4. HF general treatment process based on severity
assessment
5. HF medications contraindications, adverse
effects & additional treatment rules
6. Prognosis estimation for HF patients
HEALTHINF 2008 - International Conference on Health Informatics
310

7. Non-pharmacological management and
recommendations
8. Specific medication prescription and dosage
9. Acute decompensation of congestive heart
failure
10. Heart failure cause and CAD risk factors
An example for a rule from the diagnosis subtask is:
Diagnosis: Systolic heart failure
IF
Patient has either heart failure signs or heart failure
symptoms
AND
ECG abnormal (left bundle branch block AND
anterior Q waves)
AND
patient has (ischemic heart disease)
AND
Chest X-ray abnormal (cardiothoracic ratio > 0.5)
AND
natriuretic peptides abnormal (BNP > 100 pg/ml))
5.2 Soft Computing
Intelligent medical applications require the ability to
work with imprecise or only partially true data. The
goal is to ensure robustness and efficiency of the
decision making process in a real world
environment. Soft computing techniques including
fuzzy systems and probabilistic reasoning can be
used to solve these problems. These techniques
typically lead to relative complex systems whose
performance and final decisions are rather difficult
to predict. For the platform we have decided to solve
the problem by a) a mixture of deterministic
production rules with fuzzy output values
(consequences) and b) complex but deterministic
computation of some input values that mimic fuzzy
inputs.
5.2.1 Fuzzy Consequences from
Deterministic Rules
The approach means that we have deterministic
rules, deterministic rule inputs, and deterministic
outputs which may have different, but in advance
predefined levels of reliability or probability.
An example of the deterministic rule with fuzzy
consequence is that Heart failure is possible IF
Patient has either heart failure signs or heart failure
symptoms AND cardiothoracic ratio > 0.5. Such
rule has precisely defined conditions but the level of
the reliability of the consequence is rather low.
Higher level of reliability is the rule with the
consequence Heart failure is probable, while the
highest level is that Heart failure diagnosis is
suggested.
The advantage of this approach is that decision
making process is deterministic and consequently
relatively simple. The application of this approach is
possible for the HF platform because decisions are
directed to humans who must decide upon their
acceptability and they are not automatically
executed. Medical doctors are the only ones who can
confirm and follow these suggestions. In this
framework we do not have closed loop decisions.
Suggestions with a predefined level of reliability are
completely acceptable. Moreover, they are easier to
interpret by humans than the numerical values
produced by probabilistic reasoning and completely
fuzzy systems.
In addition to different levels of the diagnosis
reliability in which we have four levels (suggested,
probable, possible, unlikely), we use fuzzy
conclusions for the prognosis (good, worse, very
poor), for medication recommendation (suggested,
consider) etc.
5.2.2 Computation of Complex Patient
Descriptors
Some deterministic rules require inputs that describe
patient status that are difficult to define by absolute
values. Such inputs can be described as fuzzy
because the same value can in different situations
have different meaning. Good examples are all
values that should be interpreted relative to some
other, current or previous, patient characteristics and
measured values.
We avoid using fuzzy values by implementing
complex computation in the process of preparing the
inputs for decision making. This means that in this
computation we must take into account, besides
basic patient characteristic, also all other properties
that significantly determine its previous or current
status relevant for the interpretation of the basic
characteristic. For example, for the input significant
arterial drop we have to look into the complete
patient history, compute mean blood pressure
values, and then based on the current value that is
more than 30 mmHg lower conclude on significant
arterial drop.
The computation of complex descriptors can be
effectively realized inside the factual knowledge
building block. It has access to the complete set of
patient data and this enables that all data necessary
for complex computations can be acquired. If some
data can not be found in the patient record then the
final patient descriptor will have unknown value and
MEDICAL KNOWLEDGE REPRESENTATION WITHIN HEARTFAID PLATFORM
311

this will prevent the respective rule to fire. For the
sake of decision reliability, the rules should have
also simple security cut-off points, like present
systolic blood pressure below 90 mmHg in the
previous example that will fire also if data necessary
for complex computations are not available.
The main disadvantage of this approach is that
rather complex computations relevant for the
decision making process are built into fixed
programmed logic with the consequence that they
can not be changed easily. The advantages are
simplicity of procedural knowledge and the
reliability of the DSS process.
5.3 Ontological Representation of
Procedural Knowledge
Presentation of HF procedural knowledge in the
form of production rules does not mean that their
practical realization for the decision support
purposes must be in the same form. It is true that
these rules may be used to build a rule based expert
system, but these rules can also be used for
representing procedural knowledge in other forms.
For the HF platform, the integration with descriptive
knowledge presented in the HF ontology is relevant.
In this situation, an appropriate form for procedural
knowledge is SWRL (Semantic Web Rule
Language), which is a logical extension of the OWL
(Web Ontology Language).
But there is also another possibility. The unique
property of OWL is the ability to represent logical
operations between classes and between classes and
individuals using the so called concept constructors.
It enables that logical relations contained in
production rules can be presented in the ontological
form. The result is the ontology that contains both
descriptive and procedural knowledge. The
advantage of the approach is a tight connection and
conceptualized representation of complete domain
knowledge which may potentially lead to a more
intuitive representation of medical knowledge. In the
future this can also enable web based distributed
decision support, but this is not relevant for the
current platform realization.
By integrating all of the ten sets of production
rules, we have built the procedural HF ontology. It
is different than the descriptive HF ontology because
it has two root classes: "Patient" and
"Patient_caracteristics". The "Patient_characteris-
tics" class contains descriptive knowledge necessary
for logical relations in production rules. All of its
subclasses and individuals, including the class
hierarchy, are based on and can be thought of as a
subset of the HF ontology. Theoretically the
complete descriptive HF ontology could be
integrated here but this was not done because of
reasoning efficiency. So the "Patient_characteristics"
class contains only descriptive knowledge necessary
for reasoning with current version of procedural
knowledge.
The class "Patient" contains the complete
procedural knowledge. In order to be compatible
with production rules organization it has ten
subclasses, each of them representing one rule set.
They are further divided into many subclasses.
Every class with no subclasses has necessary and
sufficient conditions defined, and this is where
procedural knowledge is stored. Each of the class
definitions can be found as a rule in one of the rule
sets. Fulfilling conditions for being in the class is
perceived as a suggestion for a particular patient,
e.g. class Patient_Severity_assessment has subclass
NYHA_IV. Conditions for this class are: patient has
dyspnoea, fatigue or palpitations at rest and patient
has heart failure. The patient with these
characteristics fulfils the conditions for being in this
class.
Testing is always the significant part of the KB
development process. By presenting production
rules in the ontological form we have enabled that
the problem can be solved by developing a java-
based OWL interpreter with added concept
“negation-as-failure” into the logic semantics of
OWL. The hybrid instance checking process
obtained in that manner aims to combine the OWL
syntax with the closed world assumption semantics.
The developed Protégé plug-in integrates the
interpreter into the Protégé with a simple user
interface. In this way, the interpreter is introduced
directly into the knowledge base building facility,
which eases the process of building and maintaining
the knowledge base.
6 MEDICAL PLANS
Medical plans are textual and visual presentations of
procedures that take place after detection of some
events. Events can be any type of health disorder
including signs, symptoms, and diagnosis. The main
characteristic of medical plans is that they are event
driven and because of that they present actionable
view of the medical knowledge. This actionable
view is a special case of the more general procedural
knowledge.
HEALTHINF 2008 - International Conference on Health Informatics
312

In the Heartfaid platform medical plans are only
a middle step between experts and the guideline
modelling tools which persuade the experts to
clearly state the procedure they would normally
perform when facing a specific problem. At the
same time they enable technicians to understand it
and encode it in a machine readable form (de Clercq
et al, 2004). They are similar to medical pathways
but in contrast to medical pathways which are
designed to be used by medical doctors in order to
systemize and standardize their work, medical plans
are designed for technical people in order to better
understand medical concepts. In the HF domain we
have used medical plans as an auxiliary tool for
systemizing procedural knowledge development and
to enable some verification of the implemented
knowledge base.
The syntax of the medical plans highly
resembles the traditional workflow management.
The difference is that the medical plans will not be
executed by machines; they are written in an almost-
free graph/text form with main purpose to be fully
understandable by humans. Their main characteristic
is that they are event driven and their main
advantage is a clear systematization of the medical
procedures and interconnections among them.
Additionally, their visual presentation facilitates
understandability by medical experts.
For the heart failure platform, medical plans
describe the disorders that can occur as events to the
patient who is treated by the platform. These
disorders have assigned urgency levels which
correspond to the type of response needed from the
medical team. For example, pulmonary edema has
the highest urgency level, requiring immediate
admission to the hospital. An example of a symptom
that has a low urgency level is cough. Cough does
not require for the patient to report to the hospital,
but rather if it is persistent, he should contact his
general practitioner. The urgency levels solve the
problem of entering the appropriate medical plan in
situations when more than one triggering event
occurs. The plans of the lower urgency level can be
interrupted if another event of the higher urgency
level happens.
At the moment, the heart failure system has 38
interconnected plans for signs, symptoms and
diagnosis assessment and treatment and 15 plans for
medications prescription and dosage. Most of them
have been presented in both graphical and textual
form. Figure 2 presents the medical plan for
handling heart failure patients with increased body
temperature.
The usual way of designing medical plans is in
close resemblance to the medical doctor’s way of
thinking when handling a patient. The most common
procedure would be to ask for other symptoms in
relation to the one the patient complains about and to
do the examination and find the appropriate signs.
These other signs and symptoms can either confirm
the initial suspicion, or request that some tests
should be taken, or completely disprove the
existence of the disorder. Usually, the next thing a
medical doctor would do is to order a series of tests.
These tests can also confirm the suspicions or give
rise to a new possible diagnosis. The next course of
action is to prescribe the appropriate treatment or to
give recommendations.
6.1 Further Research Topics
In the HF project the medical plans have been
developed by technical people in the phase of
procedural knowledge development. The goals were
to demonstrate medical domain understanding and to
systemize acquired knowledge. Their significance is
in the fact that they present a middle step between
the experts and their expert knowledge and technical
people that formalize the knowledge.
Development of medical plans opened some
potentially interesting questions that might be very
relevant as further research topics. The first is
weather all types of useful medical procedural
knowledge (or at least its major part) can be
described by medical plans. If the answer is positive,
then it would be interesting to think about the
possibility to make medical plans executable directly
without their transformation to other forms (rules,
ontologies, workflows) or to try to enable their
automatic conversion without human intervention
(de Clercq et al, 2004). Based on the work and
results in the HF project, these options seem
interesting because the approach based on medical
plans as the first and potentially the only creative
part requiring human intervention, could
significantly change the traditional way of designing
procedural knowledge.
7 CONCLUSIONS
The paper presents the main results of the work
related to collection, systematization, and
formalization of the knowledge related to the heart
failure domain. The main results are: descriptive HF
MEDICAL KNOWLEDGE REPRESENTATION WITHIN HEARTFAID PLATFORM
313
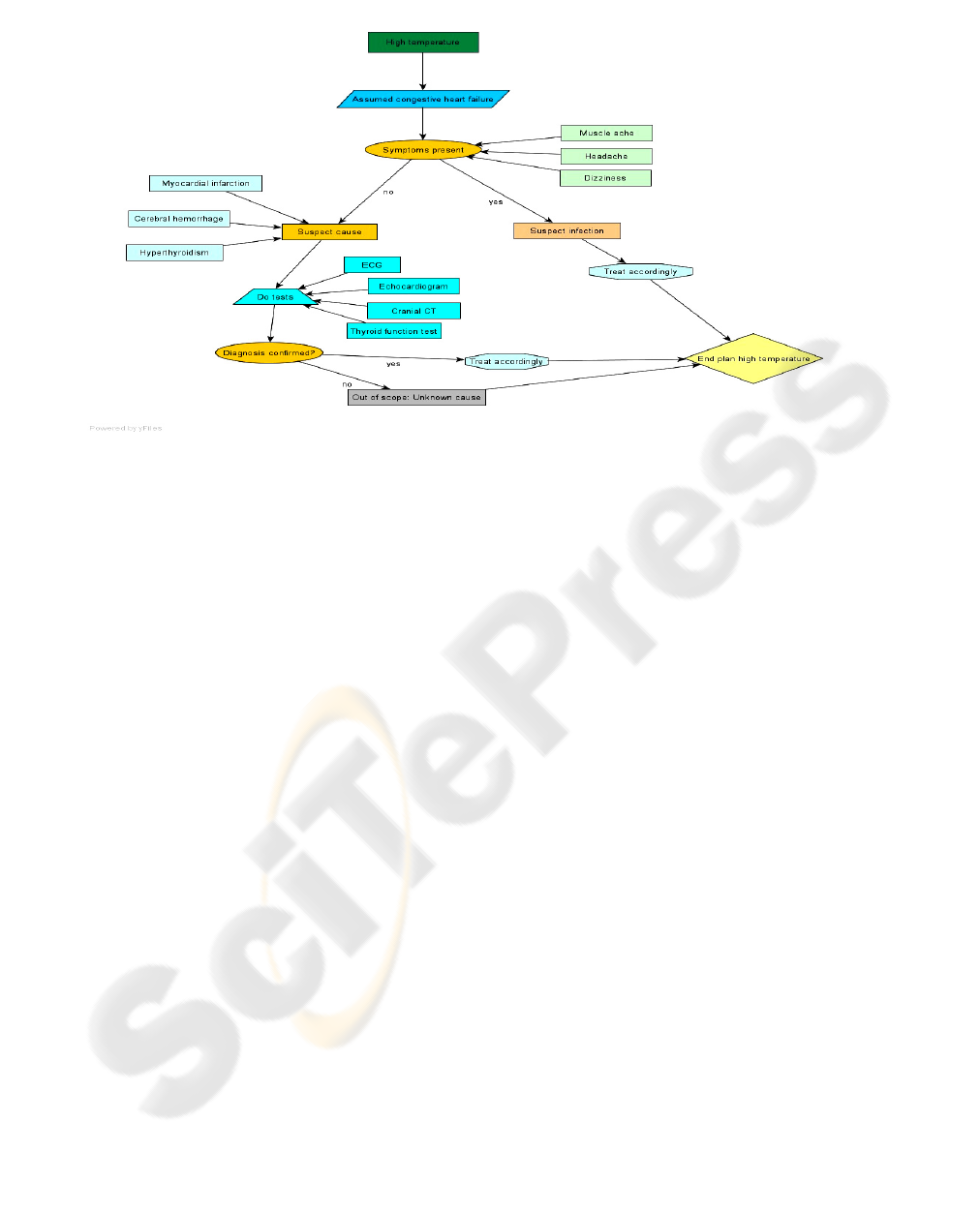
Figure 2: Medical plan for heart failure patients with increased body temperature.
ontology, procedural knowledge base, HF medical
plans, and ontological presentation of procedural
knowledge.
Although the constructed knowledge base has
been partially verified and improved by medical
doctors, the current version presents technical
formalization of medical guidelines and starting
point for Heartfaid platform implementation. It can
be expected that tests with prototypes will
demonstrate deficiencies in the form and content of
the knowledge base. By these improvements we
expect to be able to collect and formalize also the
tacit medical knowledge related to HF. Long and
detailed experimental work with the platform is the
necessary condition for the success of this process.
Moreover, even in the operational life of the
platform it can be expected that continuous
improvements in the knowledge base will be
necessary.
ACKNOWLEDGEMENTS
The work was done for EU FP6 project "Heartfaid:
A knowledge based platform of services for
supporting medical-clinical management of the heart
failure within the elderly population". It was
supported by Croatian Ministry of Science,
Education and Sport project "Machine Learning
Algorithms and Applications".
REFERENCES
Hripcsak, G., Clayton, P.D., Pryor, T.A., Haug, P.,
Wigertz, O.B., Van der lei, J., 1993. The arden syntax
for medical logic modules. Forteenth Annual
Symposium on Computer Applications in Medical
Care, 200-204. IEEE Computer Society
Press.
Peleg, M., Boxwala, A., Ogunyemi, O., 2000. GLIF3: The
Evolution of a Guideline Representation Format.
Proceedings of the American Medical Informatics
Association (AMIA) Annual Symposium. (20
Suppl):645-649. Washington DC: AMIA.
Shahar, Y., Miksch, S., Johnson, P., 1998. The Asgaard
project: a task-specific framework for the application
and critiquing of time-oriented clinical guidelines.
Artificial Intelligence in Medicine, 14(1-2), 29-51.
Springer, 1998.
de Clercq, A., Blom, J.A., Korsten, H.M., Hasman, A.,
2004. Approaches for creating computer-interpretable
guidelines that facilitate decision support. Artificial
Intelligence in Medicine 31(1), 1-27. Springer, 2004.
Sutton, D.R., Fox, J., 2003. The Syntax and Semantics of
the PROforma guideline modelling language. Journal
of the American Medical Informatics Association:
JAMIA, 2003 Sep-Oct;10(5):433-43. Hanley &
Belfus.
HEALTHINF 2008 - International Conference on Health Informatics
314
